ARID1A safeguards the canalization of the cell fate decision during osteoclastogenesis
- PMID: 39013863
- PMCID: PMC11252270
- DOI: 10.1038/s41467-024-50225-z
ARID1A safeguards the canalization of the cell fate decision during osteoclastogenesis
Abstract
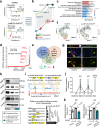
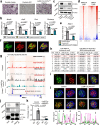
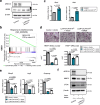
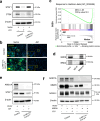
Chromatin remodeler ARID1A regulates gene transcription by modulating nucleosome positioning and chromatin accessibility. While ARID1A-mediated stage and lineage-restricted gene regulation during cell fate canalization remains unresolved. Using osteoclastogenesis as a model, we show that ARID1A transcriptionally safeguards the osteoclast (OC) fate canalization during proliferation-differentiation switching at single-cell resolution. Notably, ARID1A is indispensable for the transcriptional apparatus condensates formation with coactivator BRD4/lineage-specifying transcription factor (TF) PU.1 at Nfatc1 super-enhancer during safeguarding the OC fate canalization. Besides, the antagonist function between ARID1A-cBAF and BRD9-ncBAF complex during osteoclastogenesis has been validated with in vitro assay and compound mutant mouse model. Furthermore, the antagonistic function of ARID1A-"accelerator" and BRD9-"brake" both depend on coactivator BRD4-"clutch" during osteoclastogenesis. Overall, these results uncover sophisticated cooperation between chromatin remodeler ARID1A, coactivator, and lineage-specifying TF at super-enhancer of lineage master TF in a condensate manner, and antagonist between distinct BAF complexes in the proper and balanced cell fate canalization.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Waddington, C.H. The strategy of the genes. (Routledge, 2014).
MeSH terms
Substances
Grants and funding
- 82201004/National Natural Science Foundation of China (National Science Foundation of China)
- 81921002/National Natural Science Foundation of China (National Science Foundation of China)
- 82130027/National Natural Science Foundation of China (National Science Foundation of China)
- 82100966/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

