A multiomic atlas of the aging hippocampus reveals molecular changes in response to environmental enrichment
- PMID: 39013876
- PMCID: PMC11252340
- DOI: 10.1038/s41467-024-49608-z
A multiomic atlas of the aging hippocampus reveals molecular changes in response to environmental enrichment
Abstract
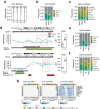
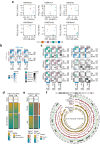
Aging involves the deterioration of organismal function, leading to the emergence of multiple pathologies. Environmental stimuli, including lifestyle, can influence the trajectory of this process and may be used as tools in the pursuit of healthy aging. To evaluate the role of epigenetic mechanisms in this context, we have generated bulk tissue and single cell multi-omic maps of the male mouse dorsal hippocampus in young and old animals exposed to environmental stimulation in the form of enriched environments. We present a molecular atlas of the aging process, highlighting two distinct axes, related to inflammation and to the dysregulation of mRNA metabolism, at the functional RNA and protein level. Additionally, we report the alteration of heterochromatin domains, including the loss of bivalent chromatin and the uncovering of a heterochromatin-switch phenomenon whereby constitutive heterochromatin loss is partially mitigated through gains in facultative heterochromatin. Notably, we observed the multi-omic reversal of a great number of aging-associated alterations in the context of environmental enrichment, which was particularly linked to glial and oligodendrocyte pathways. In conclusion, our work describes the epigenomic landscape of environmental stimulation in the context of aging and reveals how lifestyle intervention can lead to the multi-layered reversal of aging-associated decline.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
- PROYE18061FERN/Fundación Científica Asociación Española Contra el Cáncer (Scientific Foundation, Spanish Association Against Cancer)
- IDI/2018/146/Gobierno del Principado de Asturias (Government of the Principality of Asturias)
- IDI/2021/000077/Gobierno del Principado de Asturias (Government of the Principality of Asturias)
- PI18/01527/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- PI21/01067/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- COV00624/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- SGL2021-03-39/European Commission (EC)
- 202020E092/Fundación General CSIC (Fundación General Consejo Superior de Investigaciones Científicas)
- SOLAUT_00038505 SGL2103040/Fundación General CSIC (Fundación General Consejo Superior de Investigaciones Científicas)
LinkOut - more resources
Full Text Sources
Medical

