NOP2 facilitates EZH2-mediated epithelial-mesenchymal transition by enhancing EZH2 mRNA stability via m5C methylation in lung cancer progression
- PMID: 39013911
- PMCID: PMC11252406
- DOI: 10.1038/s41419-024-06899-w
NOP2 facilitates EZH2-mediated epithelial-mesenchymal transition by enhancing EZH2 mRNA stability via m5C methylation in lung cancer progression
Abstract
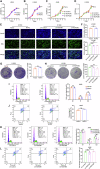
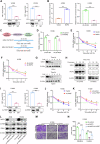
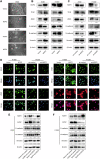
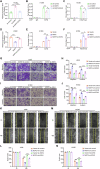
NOP2, a member of the NOL1/NOP2/SUN domain (NSUN) family, is responsible for catalyzing the posttranscriptional modification of RNA through 5-methylcytosine (m5C). Dysregulation of m5C modification has been linked to the pathogenesis of various malignant tumors. Herein, we investigated the expression of NOP2 in lung adenocarcinoma (LUAD) tissues and cells, and found that it was significantly upregulated. Moreover, lentivirus-mediated overexpression of NOP2 in vitro resulted in enhanced migration and invasion capabilities of lung cancer cells, while in vivo experiments demonstrated its ability to promote the growth and metastasis of xenograft tumors. In contrast, knockdown of NOP2 effectively inhibited the growth and metastasis of lung cancer cells. RNA-sequencing was conducted to ascertain the downstream targets of NOP2, and the findings revealed a significant upregulation in EZH2 mRNA expression upon overexpression of NOP2. Subsequent validation experiments demonstrated that NOP2 exerted an m5C-dependent influence on the stability of EZH2 mRNA. Additionally, our investigations revealed a co-regulatory relationship between NOP2 and the m5C reader protein ALYREF in modulating the stability of EZH2 mRNA. Notably, the NOP2/EZH2 axis facilitated the malignant phenotype of lung cancer cells by inducing epithelial-mesenchymal transition (EMT) both in vitro and in vivo. Mechanistically, ChIP analysis proved that EZH2 counteracted the impact of NOP2 on the occupancy capacity of EZH2 and H3K27me3 in the promoter regions of E-cadherin, a gene crucial for regulating EMT. In a word, our research highlights the significant role of NOP2 in LUAD and offers novel mechanistic insights into the NOP2/ALYREF/EZH2 axis, which holds promise as a potential target for lung cancer therapy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Collins LG, Haines C, Perkel R, Enck RE. Lung cancer: diagnosis and management. Am Fam Physician. 2007;75:56–63. - PubMed
-
- Kris MG, Gaspar LE, Chaft JE, Kennedy EB, Azzoli CG, Ellis PM, et al. Adjuvant systemic therapy and adjuvant radiation therapy for stage I to IIIA completely resected non-small-cell lung cancers: american society of clinical oncology/cancer care ontario clinical practice guideline update. J Clin Oncol. 2017;35:2960–74. doi: 10.1200/JCO.2017.72.4401. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- Grant No.232300421054/Natural Science Foundation of Henan Province (Henan Province Natural Science Foundation)
- Grant No.232300421286/Natural Science Foundation of Henan Province (Henan Province Natural Science Foundation)
- Grant No. 82173018/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

