TDP1 phosphorylation by CDK1 in mitosis promotes MUS81-dependent repair of trapped Top1-DNA covalent complexes
- PMID: 39014228
- PMCID: PMC11377750
- DOI: 10.1038/s44318-024-00169-3
TDP1 phosphorylation by CDK1 in mitosis promotes MUS81-dependent repair of trapped Top1-DNA covalent complexes
Abstract
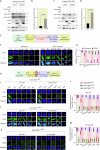
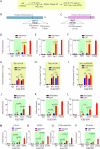
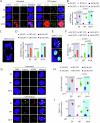
Topoisomerase 1 (Top1) controls DNA topology, relieves DNA supercoiling during replication and transcription, and is critical for mitotic progression to the G1 phase. Tyrosyl-DNA phosphodiesterase 1 (TDP1) mediates the removal of trapped Top1-DNA covalent complexes (Top1cc). Here, we identify CDK1-dependent phosphorylation of TDP1 at residue S61 during mitosis. A TDP1 variant defective for S61 phosphorylation (TDP1-S61A) is trapped on the mitotic chromosomes, triggering DNA damage and mitotic defects. Moreover, we show that Top1cc repair in mitosis occurs via a MUS81-dependent DNA repair mechanism. Replication stress induced by camptothecin or aphidicolin leads to TDP1-S61A enrichment at common fragile sites, which over-stimulates MUS81-dependent chromatid breaks, anaphase bridges, and micronuclei, ultimately culminating in the formation of 53BP1 nuclear bodies during G1 phase. Our findings provide new insights into the cell cycle-dependent regulation of TDP1 dynamics for the repair of trapped Top1-DNA covalent complexes during mitosis that prevents genomic instability following replication stress.
Keywords: CDK1; MUS81; MiDAS; TDP1; Topoisomerase 1.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

