AURKB promotes immunogenicity and immune infiltration in clear cell renal cell carcinoma
- PMID: 39014265
- PMCID: PMC11252114
- DOI: 10.1007/s12672-024-01141-7
AURKB promotes immunogenicity and immune infiltration in clear cell renal cell carcinoma
Abstract
Background: Chromatin regulators (CRs) are capable of causing epigenetic alterations, which are significant features of cancer. However, the function of CRs in controlling Clear Cell Renal Cell Carcinoma (ccRCC) is not well understood. This research aims to discover a CRs prognostic signature in ccRCC and to elucidate the roles of CRs-related genes in tumor microenvironment (TME).
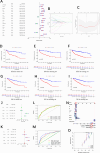
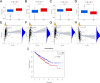
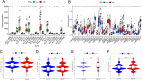
Methods: Expression profiles and relevant clinical annotations were retrieved from the Cancer Genome Atlas (TCGA) and UCSC Xena platform for progression-free survival (PFS) data. The R package "limma" was used to identify differentially expressed CRs. A predictive model based on five CRs was developed using LASSO-Cox analysis. The model's predictive power and applicability were validated using K-M curves, ROC curves, nomograms, comparisons with other models, stratified survival analyses, and validation with the ICGC cohort. GO and GSEA analyses were performed to investigate mechanisms differentiating low and high riskScore groups. Immunogenicity was assessed using Tumor Mutational Burden (TMB), immune cell infiltrations were inferred, and immunotherapy was evaluated using immunophenogram analysis and the expression patterns of human leukocyte antigen (HLA) and checkpoint genes. Differentially expressed CRs (DECRs) between low and high riskScore groups were identified using log2|FC|> 1 and FDR < 0.05. AURKB, one of the high-risk DECRs and a component of our prognostic model, was selected for further analysis.
Results: We constructed a 5 CRs signature, which demonstrated a strong capacity to predict survival and greater applicability in ccRCC. Elevated immunogenicity and immune infiltration in the high riskScore group were associated with poor prognosis. Immunotherapy was more effective in the high riskScore group, and certain chemotherapy medications, including cisplatin, docetaxel, bleomycin, and axitinib, had lower IC50 values. Our research shows that AURKB is critical for the immunogenicity and immune infiltration of the high riskScore group.
Conclusion: Our study produced a reliable prognostic prediction model using only 5 CRs. We found that AURKB promotes immunogenicity and immune infiltration. This research provides crucial support for the development of prognostic biomarkers and treatment strategies for ccRCC.
Keywords: Chromatin regulators; Immune therapy; Immunogenicity; Prognostic model; TICs; ccRCC.
© 2024. The Author(s).
Conflict of interest statement
No conflicts of interest are declared by the authors.
Figures

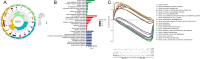



References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
