Identification and transcriptomic assessment of latent profile pediatric septic shock phenotypes
- PMID: 39014377
- PMCID: PMC11253460
- DOI: 10.1186/s13054-024-05020-z
Identification and transcriptomic assessment of latent profile pediatric septic shock phenotypes
Abstract
Background: Sepsis poses a grave threat, especially among children, but treatments are limited owing to heterogeneity among patients. We sought to test the clinical and biological relevance of pediatric septic shock subclasses identified using reproducible approaches.
Methods: We performed latent profile analyses using clinical, laboratory, and biomarker data from a prospective multi-center pediatric septic shock observational cohort to derive phenotypes and trained a support vector machine model to assign phenotypes in an internal validation set. We established the clinical relevance of phenotypes and tested for their interaction with common sepsis treatments on patient outcomes. We conducted transcriptomic analyses to delineate phenotype-specific biology and inferred underlying cell subpopulations. Finally, we compared whether latent profile phenotypes overlapped with established gene-expression endotypes and compared survival among patients based on an integrated subclassification scheme.
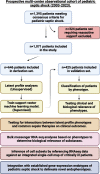
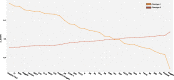
Results: Among 1071 pediatric septic shock patients requiring vasoactive support on day 1 included, we identified two phenotypes which we designated as Phenotype 1 (19.5%) and Phenotype 2 (80.5%). Membership in Phenotype 1 was associated with ~ fourfold adjusted odds of complicated course relative to Phenotype 2. Patients belonging to Phenotype 1 were characterized by relatively higher Angiopoietin-2/Tie-2 ratio, Angiopoietin-2, soluble thrombomodulin (sTM), interleukin 8 (IL-8), and intercellular adhesion molecule 1 (ICAM-1) and lower Tie-2 and Angiopoietin-1 concentrations compared to Phenotype 2. We did not identify significant interactions between phenotypes, common treatments, and clinical outcomes. Transcriptomic analysis revealed overexpression of genes implicated in the innate immune response and driven primarily by developing neutrophils among patients designated as Phenotype 1. There was no statistically significant overlap between established gene-expression endotypes, reflective of the host adaptive response, and the newly derived phenotypes, reflective of the host innate response including microvascular endothelial dysfunction. However, an integrated subclassification scheme demonstrated varying survival probabilities when comparing patient endophenotypes.
Conclusions: Our research underscores the reproducibility of latent profile analyses to identify pediatric septic shock phenotypes with high prognostic relevance. Pending validation, an integrated subclassification scheme, reflective of the different facets of the host response, holds promise to inform targeted intervention among those critically ill.
Keywords: Endotype; Phenotype; Precision medicine; Sepsis.
© 2024. The Author(s).
Conflict of interest statement
Cincinnati Children’s Hospital Medical Center (CCHMC) and the estate of the late Dr. Hector R. Wong hold patents for gene-expression-based pediatric septic shock endotypes, reflective of the host adaptive immune system. M.R.A and R.K hold a provisional patent for gene-expression-based multiple organ dysfunction syndrome (MODS) subclass identification, reflective of the host innate immune response. Inflammatix is a for-profit company focused on the development and commercialization of best-in-class host-response diagnostic tests. Y.H.B and T.E.S are employees and/or stockholders of Inflammatix Inc. P.K is a stockholder of Inflammatix Inc.
Figures



References
-
- Global report on the epidemiology and burden of sepsis: current evidence, identifying gaps and future directions. World Health Organization. 2020. p.56. Available from: https://apps.who.int/iris/bitstream/handle/10665/334216/9789240010789-en...
-
- Shah FA, Meyer NJ, Angus DC, Awdish R, Azoulay É, Calfee CS, et al. A research agenda for precision medicine in sepsis and acute respiratory distress syndrome: an Official American Thoracic Society Research Statement. Am J Respir Crit Care Med. 2021;204(8):891–901. doi: 10.1164/rccm.202108-1908ST. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01HD105939/National Institute of Child Health and Human Development
- R21GM150093/GM/NIGMS NIH HHS/United States
- R21 GM150093/GM/NIGMS NIH HHS/United States
- R01 HL149910/HL/NHLBI NIH HHS/United States
- T32 HL129970/HL/NHLBI NIH HHS/United States
- R21GM151703/GM/NIGMS NIH HHS/United States
- R35 GM126943/GM/NIGMS NIH HHS/United States
- R21 GM151703/GM/NIGMS NIH HHS/United States
- K23 GM151444/GM/NIGMS NIH HHS/United States
- K08 GM148957/GM/NIGMS NIH HHS/United States
- K23 DK119463/DK/NIDDK NIH HHS/United States
- R01 HD105939/HD/NICHD NIH HHS/United States
- R01HL149910/HL/NHLBI NIH HHS/United States
- R01 GM139967/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

