Transcription Factor 23 is an Essential Determinant of Murine Term Parturition
- PMID: 39014976
- PMCID: PMC11296541
- DOI: 10.1080/10985549.2024.2376146
Transcription Factor 23 is an Essential Determinant of Murine Term Parturition
Abstract
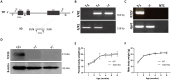
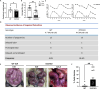
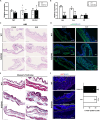
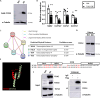
Pregnancy involving intricate tissue transformations governed by the progesterone hormone (P4). P4 signaling via P4 receptors (PRs) is vital for endometrial receptivity, decidualization, myometrial quiescence, and labor initiation. This study explored the role of TCF23 as a downstream target of PR during pregnancy. TCF23 was found to be expressed in female reproductive organs, predominantly in uterine stromal and smooth muscle cells. Tcf23 expression was high during midgestation and was specifically regulated by P4, but not estrogen. The Tcf23 knockout (KO) mouse was generated and analyzed. Female KO mice aged 4-6 months exhibited subfertility, reduced litter size, and defective parturition. Uterine histology revealed disrupted myometrial structure, altered collagen organization, and disarrayed smooth muscle sheets at the conceptus sites of KO mice. RNA-Seq analysis of KO myometrium revealed dysregulation of genes associated with cell adhesion and extracellular matrix organization. TCF23 potentially modulates TCF12 activity to mediate cell-cell adhesion and matrix modulation in smooth muscle cells. Overall, TCF23 deficiency leads to impaired myometrial remodeling, causing parturition delay and fetal demise. This study sheds light on the critical role of TCF23 as a dowstream mediator of PR in uterine remodeling, reflecting the importance of cell-cell communication and matrix dynamics in myometrial activation and parturition.
Keywords: TCF23; bHLH; dystocia; extracellular matrix; myometrium remodeling; parturition; progesterone; resorption; smooth muscle cells.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures




References
-
- Haraguchi H, Saito-Fujita T, Hirota Y, Egashira M, Matsumoto L, Matsuo M, Hiraoka T, Koga K, Yamauchi N, Fukayama M, et al. MicroRNA-200a locally attenuates progesterone signaling in the cervix, preventing embryo implantation. Mol Endocrinol. 2014;28:1108–1117. doi: 10.1210/me.2014-1097. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
