The Tumor Microbiome as a Predictor of Outcomes in Patients with Metastatic Melanoma Treated with Immune Checkpoint Inhibitors
- PMID: 39015091
- PMCID: PMC11307144
- DOI: 10.1158/2767-9764.CRC-23-0170
The Tumor Microbiome as a Predictor of Outcomes in Patients with Metastatic Melanoma Treated with Immune Checkpoint Inhibitors
Abstract
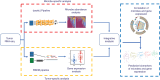
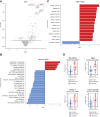
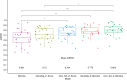
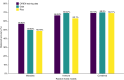
Emerging evidence supports the important role of the tumor microbiome in oncogenesis, cancer immune phenotype, cancer progression, and treatment outcomes in many malignancies. In this study, we investigated the metastatic melanoma tumor microbiome and its potential roles in association with clinical outcomes, such as survival, in patients with metastatic disease treated with immune checkpoint inhibitors (ICI). Baseline tumor samples were collected from 71 patients with metastatic melanoma before treatment with ICIs. Bulk RNA sequencing (RNA-seq) was conducted on the formalin-fixed, paraffin-embedded and fresh frozen tumor samples. Durable clinical benefit (primary clinical endpoint) following ICIs was defined as overall survival >24 months and no change to the primary drug regimen (responders). We processed RNA-seq reads to carefully identify exogenous sequences using the {exotic} tool. The age of the 71 patients with metastatic melanoma ranged from 24 to 83 years, 59% were male, and 55% survived >24 months following the initiation of ICI treatment. Exogenous taxa were identified in the tumor RNA-seq, including bacteria, fungi, and viruses. We found differences in gene expression and microbe abundances in immunotherapy-responsive versus nonresponsive tumors. Responders showed significant enrichment of bacteriophages in the phylum Uroviricota, and nonresponders showed enrichment of several bacteria, including Campylobacter jejuni. These microbes correlated with immune-related gene expression signatures. Finally, we found that models for predicting prolonged survival with immunotherapy using both microbe abundances and gene expression outperformed models using either dataset alone. Our findings warrant further investigation and potentially support therapeutic strategies to modify the tumor microbiome in order to improve treatment outcomes with ICIs.
Significance: We analyzed the tumor microbiome and interactions with genes and pathways in metastatic melanoma treated with immunotherapy and identified several microbes associated with immunotherapy response and immune-related gene expression signatures. Machine learning models that combined microbe abundances and gene expression outperformed models using either dataset alone in predicting immunotherapy responses.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
M.D. McCarter reports grants from Merck and Taiho outside the submitted work. E.A. Singer reports other support from Aura Biosciences outside the submitted work. G. Tinoco reports personal fees from Deciphera Pharmaceuticals, Daiichi Sankyo Company, and Servier Pharmaceuticals outside the submitted work. C.M. Ulrich reports nonfinancial support from Aster Insights and grants from NIH during the conduct of the study, as well as grants from NIH outside the submitted work. Y. Zakharia reports advisory board: Bristol Myers Squibb, Amgen, Roche Diagnostics, Novartis, Janssen, Eisai, Exelixis, Castle Biosciences, Genzyme Corporation, AstraZeneca, Array, Bayer, Pfizer, Clovis, EMD Serono, and Myovant; grant/research support: institution clinical trial support from NewLink Genetics, Pfizer, Exelixis, and Eisai; Data Safety and Monitoring Committee: Janssen Research and Development; and consultant honorarium: Pfizer and Novartis. D. Spakowicz reports grants from Lung Cancer Foundation of America, National Institutes on Aging, American Lung Association, Pelotonia, National Comprehensive Cancer Center Network, Oncology Nursing Foundation, Sarcoma Foundation of America, NCI, and American Cancer Society outside the submitted work. A.A. Tarhini reports grants from Bristol Myers Squibb, Merck, Novartis, Genentech-Roche, Regeneron, Sanofi-Genzyme, Pfizer, Acrotech, InflaRx, Scholar Rock, Simcha, Iovance/Clinigen, Agenus, and OncoSec and personal fees from Bristol Myers Squibb, Merck, Regeneron, Sanofi-Genzyme, ConcertAI, Teva, Instil Bio, and Pfizer outside the submitted work. No disclosures were reported by the other authors.
Figures


Update of
-
The tumor microbiome as a predictor of outcomes in patients with metastatic melanoma treated with immune checkpoint inhibitors.bioRxiv [Preprint]. 2023 May 25:2023.05.24.542123. doi: 10.1101/2023.05.24.542123. bioRxiv. 2023. Update in: Cancer Res Commun. 2024 Aug 1;4(8):1978-1990. doi: 10.1158/2767-9764.CRC-23-0170. PMID: 37292921 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

