Advances in genome sequencing and artificially induced mutation provides new avenues for cotton breeding
- PMID: 39015293
- PMCID: PMC11250495
- DOI: 10.3389/fpls.2024.1400201
Advances in genome sequencing and artificially induced mutation provides new avenues for cotton breeding
Abstract
Cotton production faces challenges in fluctuating environmental conditions due to limited genetic variation in cultivated cotton species. To enhance the genetic diversity crucial for this primary fiber crop, it is essential to augment current germplasm resources. High-throughput sequencing has significantly impacted cotton functional genomics, enabling the creation of diverse mutant libraries and the identification of mutant functional genes and new germplasm resources. Artificial mutation, established through physical or chemical methods, stands as a highly efficient strategy to enrich cotton germplasm resources, yielding stable and high-quality raw materials. In this paper, we discuss the good foundation laid by high-throughput sequencing of cotton genome for mutant identification and functional genome, and focus on the construction methods of mutant libraries and diverse sequencing strategies based on mutants. In addition, the important functional genes identified by the cotton mutant library have greatly enriched the germplasm resources and promoted the development of functional genomes. Finally, an innovative strategy for constructing a cotton CRISPR mutant library was proposed, and the possibility of high-throughput screening of cotton mutants based on a UAV phenotyping platform was discussed. The aim of this review was to expand cotton germplasm resources, mine functional genes, and develop adaptable materials in a variety of complex environments.
Keywords: cotton; diversity; germplasm resources; mutant library; sequencing.
Copyright © 2024 Wang, Abbas, He, Zhou, Cheng and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
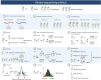
Figures




References
Publication types
LinkOut - more resources
Full Text Sources

