In vitro higher-order oligomeric assembly of the respiratory syncytial virus M2-1 protein with longer RNAs
- PMID: 39016557
- PMCID: PMC11334520
- DOI: 10.1128/jvi.01046-24
In vitro higher-order oligomeric assembly of the respiratory syncytial virus M2-1 protein with longer RNAs
Abstract
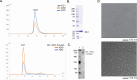
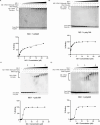
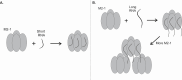
The respiratory syncytial virus (RSV) M2-1 protein is a transcriptional antitermination factor crucial for efficiently synthesizing multiple full-length viral mRNAs. During RSV infection, M2-1 exists in a complex with mRNA within cytoplasmic compartments called inclusion body-associated granules (IBAGs). Prior studies showed that M2-1 can bind along the entire length of viral mRNAs instead of just gene-end (GE) sequences, suggesting that M2-1 has more sophisticated RNA recognition and binding characteristics. Here, we analyzed the higher oligomeric complexes formed by M2-1 and RNAs in vitro using size exclusion chromatography (SEC), electrophoretic mobility shift assays (EMSA), negative stain electron microscopy (EM), and mutagenesis. We observed that the minimal RNA length for such higher oligomeric assembly is about 14 nucleotides for polyadenine sequences, and longer RNAs exhibit distinct RNA-induced binding modality to M2-1, leading to enhanced particle formation frequency and particle homogeneity as the local RNA concentration increases. We showed that particular cysteine residues of the M2-1 cysteine-cysteine-cystine-histidine (CCCH) zinc-binding motif are essential for higher oligomeric assembly. Furthermore, complexes assembled with long polyadenine sequences remain unaffected when co-incubated with ribonucleases or a zinc chelation agent. Our study provided new insights into the higher oligomeric assembly of M2-1 with longer RNA.IMPORTANCERespiratory syncytial virus (RSV) causes significant respiratory infections in infants, the elderly, and immunocompromised individuals. The virus forms specialized compartments to produce genetic material, with the M2-1 protein playing a pivotal role. M2-1 acts as an anti-terminator in viral transcription, ensuring the creation of complete viral mRNA and associating with both viral and cellular mRNA. Our research focuses on understanding M2-1's function in viral mRNA synthesis by modeling interactions in a controlled environment. This approach is crucial due to the challenges of studying these compartments in vivo. Reconstructing the system in vitro uncovers structural and biochemical aspects and reveals the potential functions of M2-1 and its homologs in related viruses. Our work may contribute to identifying targets for antiviral inhibitors and advancing RSV infection treatment.
Keywords: M2-1 protein; RNA; electrophoretic mobility shift assays (EMSA); higher-order oligomer; mutagenesis; negative stain electron microscopy (EM); respiratory syncytial virus (RSV); size exclusion chromatography (SEC).
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Respiratory syncytial virus M2-1 protein associates non-specifically with viral messenger RNA and with specific cellular messenger RNA transcripts.PLoS Pathog. 2021 May 18;17(5):e1009589. doi: 10.1371/journal.ppat.1009589. eCollection 2021 May. PLoS Pathog. 2021. PMID: 34003848 Free PMC article.
-
Functional organization of cytoplasmic inclusion bodies in cells infected by respiratory syncytial virus.Nat Commun. 2017 Sep 15;8(1):563. doi: 10.1038/s41467-017-00655-9. Nat Commun. 2017. PMID: 28916773 Free PMC article.
-
Structural Insights into the Respiratory Syncytial Virus RNA Synthesis Complexes.Viruses. 2021 May 5;13(5):834. doi: 10.3390/v13050834. Viruses. 2021. PMID: 34063087 Free PMC article. Review.
-
RSV hijacks cellular protein phosphatase 1 to regulate M2-1 phosphorylation and viral transcription.PLoS Pathog. 2018 Feb 28;14(3):e1006920. doi: 10.1371/journal.ppat.1006920. eCollection 2018 Mar. PLoS Pathog. 2018. PMID: 29489893 Free PMC article.
-
Protein-protein interactions in RSV assembly: potential targets for attenuating RSV strains.Infect Disord Drug Targets. 2012 Apr;12(2):103-9. doi: 10.2174/187152612800100125. Infect Disord Drug Targets. 2012. PMID: 22335497 Review.
Cited by
-
Regulation of respiratory syncytial virus nucleoprotein oligomerization by phosphorylation.J Biol Chem. 2025 Mar;301(3):108256. doi: 10.1016/j.jbc.2025.108256. Epub 2025 Feb 3. J Biol Chem. 2025. PMID: 39909382 Free PMC article.
References
-
- Chatzis O, Darbre S, Pasquier J, Meylan P, Manuel O, Aubert JD, Beck-Popovic M, Masouridi-Levrat S, Ansari M, Kaiser L, Posfay-Barbe KM, Asner SA. 2018. Burden of severe RSV disease among immunocompromised children and adults: a 10 year retrospective study. BMC Infect Dis 18:111. doi:10.1186/s12879-018-3002-3 - DOI - PMC - PubMed
-
- Shi T, McAllister DA, O’Brien KL, Simoes EAF, Madhi SA, Gessner BD, Polack FP, Balsells E, Acacio S, Aguayo C, et al. . 2017. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 390:946–958. doi:10.1016/S0140-6736(17)30938-8 - DOI - PMC - PubMed
-
- Lamb RA. 2007. Mononegavirales, p 1357. In Fields virology. Lippincott, Williams and Wilkins.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

