A broad survey of choanoflagellates revises the evolutionary history of the Shaker family of voltage-gated K+ channels in animals
- PMID: 39018191
- PMCID: PMC11287247
- DOI: 10.1073/pnas.2407461121
A broad survey of choanoflagellates revises the evolutionary history of the Shaker family of voltage-gated K+ channels in animals
Abstract
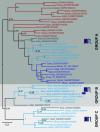
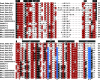
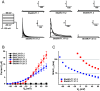
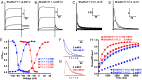
The Shaker family of voltage-gated K+ channels has been thought of as an animal-specific ion channel family that diversified in concert with nervous systems. It comprises four functionally independent gene subfamilies (Kv1-4) that encode diverse neuronal K+ currents. Comparison of animal genomes predicts that only the Kv1 subfamily was present in the animal common ancestor. Here, we show that some choanoflagellates, the closest protozoan sister lineage to animals, also have Shaker family K+ channels. Choanoflagellate Shaker family channels are surprisingly most closely related to the animal Kv2-4 subfamilies which were believed to have evolved only after the divergence of ctenophores and sponges from cnidarians and bilaterians. Structural modeling predicts that the choanoflagellate channels share a T1 Zn2+ binding site with Kv2-4 channels that is absent in Kv1 channels. We functionally expressed three Shakers from Salpingoeca helianthica (SheliKvT1.1-3) in Xenopus oocytes. SheliKvT1.1-3 function only in two heteromultimeric combinations (SheliKvT1.1/1.2 and SheliKvT1.1/1.3) and encode fast N-type inactivating K+ channels with distinct voltage dependence that are most similar to the widespread animal Kv1-encoded A-type Shakers. Structural modeling of the T1 assembly domain supports a preference for heteromeric assembly in a 2:2 stoichiometry. These results push the origin of the Shaker family back into a common ancestor of metazoans and choanoflagellates. They also suggest that the animal common ancestor had at least two distinct molecular lineages of Shaker channels, a Kv1 subfamily lineage predicted from comparison of animal genomes and a Kv2-4 lineage predicted from comparison of animals and choanoflagellates.
Keywords: Shaker; choanoflagellate; potassium channel; voltage-gated.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




Similar articles
-
Ctenophores and parahoxozoans independently evolved functionally diverse voltage-gated K+ channels.J Gen Physiol. 2025 May 5;157(3):e202413740. doi: 10.1085/jgp.202413740. Epub 2025 Mar 18. J Gen Physiol. 2025. PMID: 40100064
-
Major diversification of voltage-gated K+ channels occurred in ancestral parahoxozoans.Proc Natl Acad Sci U S A. 2015 Mar 3;112(9):E1010-9. doi: 10.1073/pnas.1422941112. Epub 2015 Feb 17. Proc Natl Acad Sci U S A. 2015. PMID: 25691740 Free PMC article.
-
Expanded functional diversity of shaker K(+) channels in cnidarians is driven by gene expansion.PLoS One. 2012;7(12):e51366. doi: 10.1371/journal.pone.0051366. Epub 2012 Dec 10. PLoS One. 2012. PMID: 23251506 Free PMC article.
-
Evolution and Structural Characteristics of Plant Voltage-Gated K+ Channels.Plant Cell. 2018 Dec;30(12):2898-2909. doi: 10.1105/tpc.18.00523. Epub 2018 Nov 1. Plant Cell. 2018. PMID: 30389753 Free PMC article. Review.
-
Involvement of the S4-S5 linker and the C-linker domain regions to voltage-gating in plant Shaker channels: comparison with animal HCN and Kv channels.Plant Signal Behav. 2014;9(10):e972892. doi: 10.4161/15592316.2014.972892. Plant Signal Behav. 2014. PMID: 25482770 Free PMC article. Review.
Cited by
-
How the Blind Watchmaker messed around with potassium channels.J Gen Physiol. 2025 May 5;157(3):e202513783. doi: 10.1085/jgp.202513783. Epub 2025 Mar 19. J Gen Physiol. 2025. PMID: 40105910
-
A Kv2 inhibitor combination reveals native neuronal conductances consistent with Kv2/KvS heteromers.Elife. 2025 May 27;13:RP99410. doi: 10.7554/eLife.99410. Elife. 2025. PMID: 40423692 Free PMC article.
-
Structural basis of fast N-type inactivation in Kv channels.Nature. 2025 Aug 6. doi: 10.1038/s41586-025-09339-7. Online ahead of print. Nature. 2025. PMID: 40770100
-
Ctenophores and parahoxozoans independently evolved functionally diverse voltage-gated K+ channels.J Gen Physiol. 2025 May 5;157(3):e202413740. doi: 10.1085/jgp.202413740. Epub 2025 Mar 18. J Gen Physiol. 2025. PMID: 40100064
References
-
- Papazian D. M., Schwarz T. L., Tempel B. L., Jan Y. N., Jan L. Y., Cloning of genomic and complementary DNA from Shaker, a putative potassium channel gene from Drosophila. Science 237, 749–753 (1987). - PubMed
-
- Kamb A., Iverson L. E., Tanouye M. A., Molecular characterization of Shaker, a Drosophila gene that encodes a potassium channel. Cell 50, 405–413 (1987). - PubMed
-
- Butler A., Wei A. G., Baker K., Salkoff L., A family of putative potassium channel genes in Drosophila. Science 243, 943–947 (1989). - PubMed
-
- Salkoff L., et al. , An essential “set” of K+ channels conserved in flies, mice and humans. Trends Neurosci. 15, 161–166 (1992). - PubMed
-
- Wei A., Jegla T., Salkoff L., Eight potassium channel families revealed by the C. elegans genome project. Neuropharmacology 35, 805–829 (1996). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

