Chondroitin sulfate proteoglycan 4 increases invasion of recessive dystrophic epidermolysis bullosa-associated cutaneous squamous cell carcinoma by modifying transforming growth factor-β signalling
- PMID: 39018437
- PMCID: PMC11663483
- DOI: 10.1093/bjd/ljae295
Chondroitin sulfate proteoglycan 4 increases invasion of recessive dystrophic epidermolysis bullosa-associated cutaneous squamous cell carcinoma by modifying transforming growth factor-β signalling
Abstract
Background: Recessive dystrophic epidermolysis bullosa (RDEB) is a rare genetic skin-blistering disorder that often progresses to metastatic cutaneous squamous cell carcinoma (cSCC) at chronic wound sites. Chondroitin sulfate proteoglycan 4 (CSPG4) is a cell-surface proteoglycan that is an oncoantigen in multiple malignancies, where it modulates oncogenic signalling, drives epithelial-to-mesenchymal transition (EMT) and enables cell motility.
Objectives: To evaluate CSPG4 expression and function in RDEB cSCC.
Methods: RDEB cSCC cell lines were used to assess CSPG4-dependent changes in invasive potential, transforming growth factor (TGF)-β1-stimulated signal activation and clinically relevant cytopathology metrics in an in vitro full-thickness tumour model. CSPG4 expression in RDEB cSCC and non-RDEB cSCC tumours was analysed via immunohistochemistry and single-cell RNA sequencing (scRNA-Seq), respectively.
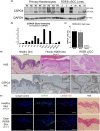
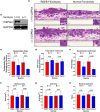
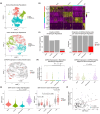
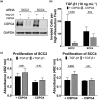
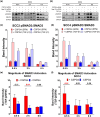
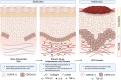
Results: Inhibiting CSPG4 expression reduced invasive potential in multiple RDEB cSCC cell lines and altered membrane-proximal TGF-β signal activation via changes in SMAD3 phosphorylation. CSPG4 expression was uniformly localized to basal layer keratinocytes in fibrotic RDEB skin and tumour cells at the tumour-stroma interface at the invasive front in RDEB cSCC tumours in vivo. Analysis of published scRNA-Seq data revealed that CSPG4 expression was correlated with an enhanced EMT transcriptomic signature in cells at the tumour-stroma interface of non-RDEB cSCC tumours. Cytopathological metrics, for example nucleus : cell area ratio, were influenced by CSPG4 expression in in vitro tumour models.
Conclusions: We determined that CSPG4 expression in RDEB cSCC cell lines enhanced the invasive potential of tumours. Mechanistically, CSPG4 was found to enhance membrane-proximal TGF-β-stimulated signalling via SMAD3, which is a key mediator of EMT in RDEB cSCC. The implication of these studies is that CSPG4 may represent a therapeutic target that can be leveraged for the clinical management of patients with RDEB cSCC.
Plain language summary
Recessive dystrophic epidermolysis bullosa (or ‘RDEB’) is a rare skin disease that affects 3 in 1 million children worldwide. People with RDEB lack an essential protein that allows the skin to resist tearing. This results in widespread blistering on the body and chronic wounds. People with RDEB can develop a lethal form of ‘metastatic’ skin cancer (where cancer cells spread to other parts of the body). However, before a cancer becomes metastatic, a cell must ‘invade’ its original tissue for it to access other parts of the body. This USA-based study investigated a protein called ‘CSPG4’. This protein is present on the surfaces of cancer cells but not in most normal tissues. Our aim was to look at whether CSPG4 enhances the invasion of skin cancer associated with RDEB. This mechanism could be targeted by anti-cancer treatments. We used skin cancer cells from people with RDEB to demonstrate that CSPG4 increases the invasive ability of cancer cells. It does this by changing how the cells respond to a signal (called ‘TGF-β’) that is present in high levels in RDEB skin. We used cancer tissue from people with RDEB to show that CSPG4 is produced by cells at the edges of tumours. These cells are responsible for invading the surrounding normal tissue. Our findings suggest that CSPG4 could be a therapeutic target. Targeting this protein may limit the ability of RDEB-associated skin cancer to invade and metastasize.
© The Author(s) 2024. Published by Oxford University Press on behalf of British Association of Dermatologists. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflicts of interest The authors declare no conflicts of interest.
Figures
References
-
- Has C, Bauer JW, Bodemer C et al. Consensus reclassification of inherited epidermolysis bullosa and other disorders with skin fragility. Br J Dermatol 2020; 183:614–27. - PubMed
-
- Kim M, Li M, Intong-Wheeler L et al. Epidemiology and outcome of squamous cell carcinoma in epidermolysis bullosa in Australia and New Zealand. Acta Derm Venereol 2018; 98:70–6. - PubMed
-
- Fine J-D, Johnson LB, Weiner M et al. Epidermolysis bullosa and the risk of life-threatening cancers: the National EB Registry experience, 1986–2006. J Am Acad Dermatol 2009; 60:203–11. - PubMed
-
- Mellerio JE, Robertson SJ, Bernardis C et al. Management of cutaneous squamous cell carcinoma in patients with epidermolysis bullosa: best clinical practice guidelines. Br J Dermatol 2016; 174:56–67. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

