Dysregulation of FLVCR1a-dependent mitochondrial calcium handling in neural progenitors causes congenital hydrocephalus
- PMID: 39019006
- PMCID: PMC11293339
- DOI: 10.1016/j.xcrm.2024.101647
Dysregulation of FLVCR1a-dependent mitochondrial calcium handling in neural progenitors causes congenital hydrocephalus
Abstract
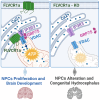
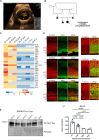
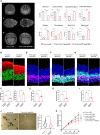
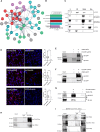
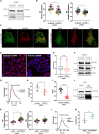
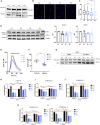
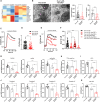
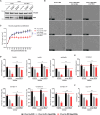
Congenital hydrocephalus (CH), occurring in approximately 1/1,000 live births, represents an important clinical challenge due to the limited knowledge of underlying molecular mechanisms. The discovery of novel CH genes is thus essential to shed light on the intricate processes responsible for ventricular dilatation in CH. Here, we identify FLVCR1 (feline leukemia virus subgroup C receptor 1) as a gene responsible for a severe form of CH in humans and mice. Mechanistically, our data reveal that the full-length isoform encoded by the FLVCR1 gene, FLVCR1a, interacts with the IP3R3-VDAC complex located on mitochondria-associated membranes (MAMs) that controls mitochondrial calcium handling. Loss of Flvcr1a in mouse neural progenitor cells (NPCs) affects mitochondrial calcium levels and energy metabolism, leading to defective cortical neurogenesis and brain ventricle enlargement. These data point to defective NPCs calcium handling and metabolic activity as one of the pathogenetic mechanisms driving CH.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests E.T., V.F., D.Chiabrando, S.P., F.B., and A.L.A. are inventors in a patent filed by the University of Torino, not related to the research reported here.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

