Replication timing alterations are associated with mutation acquisition during breast and lung cancer evolution
- PMID: 39019871
- PMCID: PMC11255325
- DOI: 10.1038/s41467-024-50107-4
Replication timing alterations are associated with mutation acquisition during breast and lung cancer evolution
Abstract
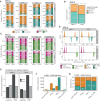
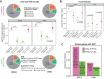
During each cell cycle, the process of DNA replication timing is tightly regulated to ensure the accurate duplication of the genome. The extent and significance of alterations in this process during malignant transformation have not been extensively explored. Here, we assess the impact of altered replication timing (ART) on cancer evolution by analysing replication-timing sequencing of cancer and normal cell lines and 952 whole-genome sequenced lung and breast tumours. We find that 6%-18% of the cancer genome exhibits ART, with regions with a change from early to late replication displaying an increased mutation rate and distinct mutational signatures. Whereas regions changing from late to early replication contain genes with increased expression and present a preponderance of APOBEC3-mediated mutation clusters and associated driver mutations. We demonstrate that ART occurs relatively early during cancer evolution and that ART may have a stronger correlation with mutation acquisition than alterations in chromatin structure.
© 2024. The Author(s).
Conflict of interest statement
N.M. has stock options in and has consulted for Achilles Therapeutics and holds a European patent in determining HLA LOH (PCT/GB2018/052004) and is a co-inventor to a patent to identifying responders to cancer treatment (PCT/GB2018/051912). N.K. acknowledges grant support from AstraZeneca. C.S. acknowledges grant support from AstraZeneca, Boehringer-Ingelheim, Bristol Myers Squibb, Pfizer, Roche-Ventana, Invitae (previously Archer Dx Inc. - collaboration in minimal residual disease sequencing technologies), and Ono Pharmaceutical. He is an AstraZeneca Advisory Board member and Chief Investigator for the AZ MeRmaiD 1 and 2 clinical trials and is also chief investigator of the NHS Galleri trial. He has consulted for Achilles Therapeutics, Amgen, AstraZeneca, Pfizer, Novartis, GlaxoSmithKline, MSD, Bristol Myers Squibb, Illumina, Genentech, Roche-Ventana, GRAIL, Medicxi, Metabomed, Bicycle Therapeutics, Roche Innovation Centre Shanghai, and the Sarah Cannon Research Institute, C.S. had stock options in Apogen Biotechnologies and GRAIL until June 2021, and currently has stock options in Epic Bioscience, Bicycle Therapeutics, and has stock options and is co-founder of Achilles Therapeutics. C.S. holds patents relating to assay technology to detect tumour recurrence (PCT/GB2017/053289); to targeting neoantigens (PCT/EP2016/059401), identifying patent response to immune checkpoint blockade (PCT/EP2016/071471), determining HLA LOH (PCT/GB2018/052004), predicting survival rates of patients with cancer (PCT/GB2020/050221), identifying patients who respond to cancer treatment (PCT/GB2018/051912), US patent relating to detecting tumour mutations (PCT/US2017/28013), methods for lung cancer detection (US20190106751A1) and both a European and US patent related to identifying insertion/deletion mutation targets (PCT/GB2018/051892). The remaining authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

