Host-derived protein profiles of human neonatal meconium across gestational ages
- PMID: 39019879
- PMCID: PMC11255260
- DOI: 10.1038/s41467-024-49805-w
Host-derived protein profiles of human neonatal meconium across gestational ages
Erratum in
-
Author Correction: Host-derived protein profiles of human neonatal meconium across gestational ages.Nat Commun. 2025 Jun 25;16(1):5398. doi: 10.1038/s41467-025-61278-z. Nat Commun. 2025. PMID: 40562747 Free PMC article. No abstract available.
Abstract
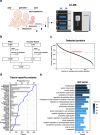
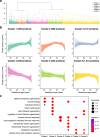
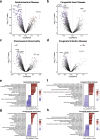
Meconium, a non-invasive biomaterial reflecting prenatal substance accumulation, could provide valuable insights into neonatal health. However, the comprehensive protein profile of meconium across gestational ages remains unclear. Here, we conducted an extensive proteomic analysis of first meconium from 259 newborns across varied gestational ages to delineate protein composition and elucidate its relevance to neonatal diseases. The first meconium samples were collected, with the majority obtained before feeding, and the mean time for the first meconium passage from the anus was 11.9 ± 9.47 h. Our analysis revealed 5370 host-derived meconium proteins, which varied depending on sex and gestational age. Specifically, meconium from preterm infants exhibited elevated concentrations of proteins associated with the extracellular matrix. Additionally, the protein profiles of meconium also exhibited unique variations depending on both specific diseases, including gastrointestinal diseases, congenital heart diseases, and maternal conditions. Furthermore, we developed a machine learning model to predict gestational ages using meconium proteins. Our model suggests that newborns with gastrointestinal diseases and congenital heart diseases may have immature gastrointestinal systems. These findings highlight the intricate relationship between clinical parameters and meconium protein composition, offering potential for a novel approach to assess neonatal gastrointestinal health.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

