miR-29a-3p orchestrates key signaling pathways for enhanced migration of human mesenchymal stem cells
- PMID: 39020373
- PMCID: PMC11256664
- DOI: 10.1186/s12964-024-01737-0
miR-29a-3p orchestrates key signaling pathways for enhanced migration of human mesenchymal stem cells
Abstract
Background: The homing of human mesenchymal stem cells (hMSCs) is crucial for their therapeutic efficacy and is characterized by the orchestrated regulation of multiple signaling modules. However, the principal upstream regulators that synchronize these signaling pathways and their mechanisms during cellular migration remain largely unexplored.
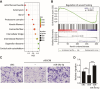
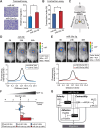
Methods: miR-29a-3p was exogenously expressed in either wild-type or DiGeorge syndrome critical region 8 (DGCR8) knockdown hMSCs. Multiple pathway components were analyzed using Western blotting, immunohistochemistry, and real-time quantitative PCR. hMSC migration was assessed both in vitro and in vivo through wound healing, Transwell, contraction, and in vivo migration assays. Extensive bioinformatic analyses using gene set enrichment analysis and Ingenuity pathway analysis identified enriched pathways, upstream regulators, and downstream targets.
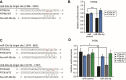
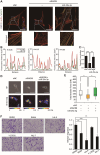
Results: The global depletion of microRNAs (miRNAs) due to DGCR8 gene silencing, a critical component of miRNA biogenesis, significantly impaired hMSC migration. The bioinformatics analysis identified miR-29a-3p as a pivotal upstream regulator. Its overexpression in DGCR8-knockdown hMSCs markedly improved their migration capabilities. Our data demonstrate that miR-29a-3p enhances cell migration by directly inhibiting two key phosphatases: protein tyrosine phosphatase receptor type kappa (PTPRK) and phosphatase and tensin homolog (PTEN). The ectopic expression of miR-29a-3p stabilized the polarization of the Golgi apparatus and actin cytoskeleton during wound healing. It also altered actomyosin contractility and cellular traction forces by changing the distribution and phosphorylation of myosin light chain 2. Additionally, it regulated focal adhesions by modulating the levels of PTPRK and paxillin. In immunocompromised mice, the migration of hMSCs overexpressing miR-29a-3p toward a chemoattractant significantly increased.
Conclusions: Our findings identify miR-29a-3p as a key upstream regulator that governs hMSC migration. Specifically, it was found to modulate principal signaling pathways, including polarization, actin cytoskeleton, contractility, and adhesion, both in vitro and in vivo, thereby reinforcing migration regulatory circuits.
Keywords: Cellular migration; Human mesenchymal stem cells; PTEN; PTPRK; miR-29a-3p.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

