Protein-free media for cardiac differentiation of hPSCs in 2000 mL suspension culture
- PMID: 39020441
- PMCID: PMC11256493
- DOI: 10.1186/s13287-024-03826-w
Protein-free media for cardiac differentiation of hPSCs in 2000 mL suspension culture
Abstract
Background: Commonly used media for the differentiation of human pluripotent stem cells into cardiomyocytes (hPSC-CMs) contain high concentrations of proteins, in particular albumin, which is prone to quality variations and presents a substantial cost factor, hampering the clinical translation of in vitro-generated cardiomyocytes for heart repair. To overcome these limitations, we have developed chemically defined, entirely protein-free media based on RPMI, supplemented with L-ascorbic acid 2-phosphate (AA-2P) and either the non-ionic surfactant Pluronic F-68 or a specific polyvinyl alcohol (PVA).
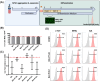
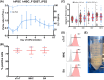
Methods and results: Both media compositions enable the efficient, directed differentiation of embryonic and induced hPSCs, matching the cell yields and cardiomyocyte purity ranging from 85 to 99% achieved with the widely used protein-based CDM3 medium. The protein-free differentiation approach was readily up-scaled to a 2000 mL process scale in a fully controlled stirred tank bioreactor in suspension culture, producing > 1.3 × 109 cardiomyocytes in a single process run. Transcriptome analysis, flow cytometry, electrophysiology, and contractile force measurements revealed that the mass-produced cardiomyocytes differentiated in protein-free medium exhibit the expected ventricular-like properties equivalent to the well-established characteristics of CDM3-control cells.
Conclusions: This study promotes the robustness and upscaling of the cardiomyogenic differentiation process, substantially reduces media costs, and provides an important step toward the clinical translation of hPSC-CMs for heart regeneration.
Keywords: Bioreactor; Cardiomyocytes; Protein-free differentiation media; hPSC.
© 2024. The Author(s).
Conflict of interest statement
F.M. and W.T. are employees of Evotec. The other authors declare no potential competing interests.
Figures



References
-
- Chong JJH, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, Gantz JA, Fugate JA, Muskheli V, Gough GM, Vogel KW, Astley CA, Hotchkiss CE, Baldessari A, Pabon L, et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510:273–277. doi: 10.1038/nature13233. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

