Discovering cancer stem-like molecule, nuclear factor I X, using spatial transcriptome in gastric cancer
- PMID: 39021298
- PMCID: PMC11462935
- DOI: 10.1111/cas.16288
Discovering cancer stem-like molecule, nuclear factor I X, using spatial transcriptome in gastric cancer
Abstract
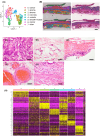
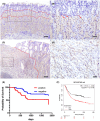
Gastric cancer (GC) is characterized by significant intratumoral heterogeneity, and stem cells are promising therapeutic targets. Despite advancements in spatial transcriptome analyses, unexplored targets for addressing cancer stemness remain unknown. This study aimed to identify Nuclear Factor IX (NFIX) as a critical regulator of cancer stemness in GC and evaluate its clinicopathological significance and function. Spatial transcriptome analysis of GC was conducted. The correlation between NFIX expression, clinicopathological factors, and prognosis was assessed using immunostaining in 127 GC cases. Functional analyses of cancer cell lines validated these findings. Spatial transcriptome analysis stratified GC tissues based on genetic profiles, identified CSC-like cells, and further refined the classification to identify and highlight the significance of NFIX, as validated by Monocle 3 and CytoTRACE analyses. Knockdown experiments in cancer cell lines have demonstrated the involvement of NFIX in cancer cell proliferation and kinase activity. This study underscores the role of spatial transcriptome analysis in refining GC tissue classification and identifying therapeutic targets, highlighting NFIX as a pivotal factor. NFIX expression is correlated with poor prognosis and drives GC progression, suggesting its potential as a novel therapeutic target for personalized GC therapies.
Keywords: biomarker; gastric cancer; gastric mucin phenotype; nuclear factor IX; spatial transcriptome.
© 2024 The Author(s). Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
Wataru Yasui is an editorial board member of
Figures




References
-
- Thuler LCS. The epidemiology of stomach cancer. In: Morgado‐Diaz JA, ed. Gastrointestinal Cancers; Brisbane (AU): Exon Publications. 2022. https://www.ncbi.nlm.nih.gov/books/NBK585996/ - PubMed
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17‐48. - PubMed
-
- Oue N, Sentani K, Sakamoto N, Uraoka N, Yasui W. Molecular carcinogenesis of gastric cancer: Lauren classification, mucin phenotype expression, and cancer stem cells. Int J Clin Oncol. 2019;24:771‐778. - PubMed
-
- Ishikawa A, Sakamoto N, Honma R, et al. Annexin A10 is involved in the induction of pancreatic duodenal homeobox‐1 in gastric cancer tissue, cells and organoids. Oncol Rep. 2020;43:581‐590. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

