DNA cytosine methyltransferases differentially regulate genome-wide hypermutation and interhomolog recombination in Trichoderma reesei meiosis
- PMID: 39021337
- PMCID: PMC11381340
- DOI: 10.1093/nar/gkae611
DNA cytosine methyltransferases differentially regulate genome-wide hypermutation and interhomolog recombination in Trichoderma reesei meiosis
Abstract
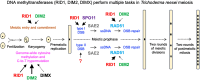
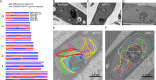
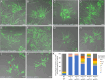
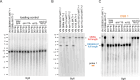
Trichoderma reesei is an economically important enzyme producer with several unique meiotic features. spo11, the initiator of meiotic double-strand breaks (DSBs) in most sexual eukaryotes, is dispensable for T. reesei meiosis. T. reesei lacks the meiosis-specific recombinase Dmc1. Rad51 and Sae2, the activator of the Mre11 endonuclease complex, promote DSB repair and chromosome synapsis in wild-type and spo11Δ meiosis. DNA methyltransferases (DNMTs) perform multiple tasks in meiosis. Three DNMT genes (rid1, dim2 and dimX) differentially regulate genome-wide cytosine methylation and C:G-to-T:A hypermutations in different chromosomal regions. We have identified two types of DSBs: type I DSBs require spo11 or rid1 for initiation, whereas type II DSBs do not rely on spo11 and rid1 for initiation. rid1 (but not dim2) is essential for Rad51-mediated DSB repair and normal meiosis. rid1 and rad51 exhibit a locus heterogeneity (LH) relationship, in which LH-associated proteins often regulate interconnectivity in protein interaction networks. This LH relationship can be suppressed by deleting dim2 in a haploid rid1Δ (but not rad51Δ) parental strain, indicating that dim2 and rid1 share a redundant function that acts earlier than rad51 during early meiosis. In conclusion, our studies provide the first evidence of the involvement of DNMTs during meiotic initiation and recombination.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






References
-
- Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018; 19:81–92. - PubMed
-
- Keeney S., Giroux C.N., Kleckner N.. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997; 88:375–384. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

