Survival prediction landscape: an in-depth systematic literature review on activities, methods, tools, diseases, and databases
- PMID: 39021434
- PMCID: PMC11252047
- DOI: 10.3389/frai.2024.1428501
Survival prediction landscape: an in-depth systematic literature review on activities, methods, tools, diseases, and databases
Abstract
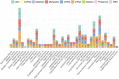
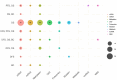
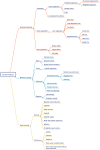
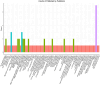
Survival prediction integrates patient-specific molecular information and clinical signatures to forecast the anticipated time of an event, such as recurrence, death, or disease progression. Survival prediction proves valuable in guiding treatment decisions, optimizing resource allocation, and interventions of precision medicine. The wide range of diseases, the existence of various variants within the same disease, and the reliance on available data necessitate disease-specific computational survival predictors. The widespread adoption of artificial intelligence (AI) methods in crafting survival predictors has undoubtedly revolutionized this field. However, the ever-increasing demand for more sophisticated and effective prediction models necessitates the continued creation of innovative advancements. To catalyze these advancements, it is crucial to bring existing survival predictors knowledge and insights into a centralized platform. The paper in hand thoroughly examines 23 existing review studies and provides a concise overview of their scope and limitations. Focusing on a comprehensive set of 90 most recent survival predictors across 44 diverse diseases, it delves into insights of diverse types of methods that are used in the development of disease-specific predictors. This exhaustive analysis encompasses the utilized data modalities along with a detailed analysis of subsets of clinical features, feature engineering methods, and the specific statistical, machine or deep learning approaches that have been employed. It also provides insights about survival prediction data sources, open-source predictors, and survival prediction frameworks.
Keywords: artificial intelligence; cancer; machine learning; multiomics; survival prediction.
Copyright © 2024 Abbasi, Asim, Ahmed, Vollmer and Dengel.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
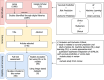
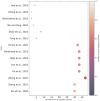
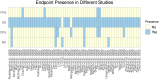
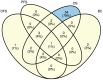
Figures

References
-
- Abdelhamid S., Scioscia J., Li S., Das J., Rahman S., Bonaroti J. W., et al. (2022). Multi-omic admission-based biomarkers predict 30-day survival and persistent critical illness in trauma patients after injury. J. Am. Coll. Surg. 235:S95. 10.1097/01.XCS.0000896540.67938.36 - DOI
Publication types
LinkOut - more resources
Full Text Sources

