Genetic specialization of key bifidobacterial phylotypes in multiple mother-infant dyad cohorts from geographically isolated populations
- PMID: 39021621
- PMCID: PMC11251887
- DOI: 10.3389/fmicb.2024.1399743
Genetic specialization of key bifidobacterial phylotypes in multiple mother-infant dyad cohorts from geographically isolated populations
Abstract
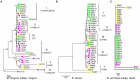
Little has been known about symbiotic relationships and host specificity for symbionts in the human gut microbiome so far. Bifidobacteria are a paragon of the symbiotic bacteria biota in the human gut. In this study, we characterized the population genetic structure of three bifidobacteria species from 58 healthy mother-infant pairs of three ethnic groups in China, geographically isolated, by Rep-PCR, multi-locus sequence analysis (MLSA), and in vitro carbohydrate utilization. One hundred strains tested were incorporated into 50 sequence types (STs), of which 29 STs, 17 STs, and 4 STs belong to B. longum subsp. longum, B. breve, and B. animalis subsp. lactis, respectively. The conspecific strains from the same mother-child pair were genetically very similar, supporting the vertical transmission of Bifidobacterium phylotypes from mother to offspring. In particular, results based on allele profiles and phylogeny showed that B. longum subsp. longum and B. breve exhibited considerable intraspecies genetic heterogeneity across three ethnic groups, and strains were clustered into ethnicity-specific lineages. Yet almost all strains of B. animalis subsp. lactis were incorporated into the same phylogenetic clade, regardless of ethnic origin. Our findings support the hypothesis of co-evolution between human gut symbionts and their respective populations, which is closely linked to the lifestyle of specific bacterial lineages. Hence, the natural and evolutionary history of Bifidobacterium species would be an additional consideration when selecting bifidobacterial strains for industrial and therapeutic applications.
Keywords: Bifidobacterium; ethnicity; mother–infant dyad; multilocus sequence typing; probiotic.
Copyright © 2024 Aihetanmu, Liang, Zhang, Luo, Zhang, Huang, Tian, Sun and Ni.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Evaluation of Bacterial Diversity and Evolutionary Dynamics of Gut Bifidobacterium longum Isolates Obtained from Older Individuals in Hubei Province, China.Microbiol Spectr. 2022 Feb 23;10(1):e0144221. doi: 10.1128/spectrum.01442-21. Epub 2022 Jan 19. Microbiol Spectr. 2022. PMID: 35044201 Free PMC article.
-
Galacto- and Fructo-oligosaccharides Utilized for Growth by Cocultures of Bifidobacterial Species Characteristic of the Infant Gut.Appl Environ Microbiol. 2020 May 19;86(11):e00214-20. doi: 10.1128/AEM.00214-20. Print 2020 May 19. Appl Environ Microbiol. 2020. PMID: 32220841 Free PMC article.
-
Genotyping and plant-derived glycan utilization analysis of Bifidobacterium strains from mother-infant pairs.BMC Microbiol. 2020 Sep 10;20(1):277. doi: 10.1186/s12866-020-01962-w. BMC Microbiol. 2020. PMID: 32912151 Free PMC article.
-
Plant Glycan Metabolism by Bifidobacteria.Front Microbiol. 2021 Feb 4;12:609418. doi: 10.3389/fmicb.2021.609418. eCollection 2021. Front Microbiol. 2021. PMID: 33613480 Free PMC article. Review.
-
Probiotics in the New Era of Human Milk Oligosaccharides (HMOs): HMO Utilization and Beneficial Effects of Bifidobacterium longum subsp. infantis M-63 on Infant Health.Microorganisms. 2024 May 17;12(5):1014. doi: 10.3390/microorganisms12051014. Microorganisms. 2024. PMID: 38792843 Free PMC article. Review.
References
-
- Barratt M. J., Nuzhat S., Ahsan K., Frese S. A., Arzamasov A. A., Sarker S. A., et al. . (2022). Bifidobacterium infantis treatment promotes weight gain in Bangladeshi infants with severe acute malnutrition. Sci. Transl. Med. 14:eabk1107. doi: 10.1126/scitranslmed.abk1107, PMID: - DOI - PMC - PubMed
-
- Bogaert D., Van Beveren G. J., De Koff E. M., Lusarreta Parga P., Balcazar Lopez C. E., Koppensteiner L., et al. . (2023). Mother-to-infant microbiota transmission and infant microbiota development across multiple body sites. Cell Host Microbe 31:e446, 447–460.e6. doi: 10.1016/j.chom.2023.01.018, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources

