Isolation and characterization of novel acetogenic strains of the genera Terrisporobacter and Acetoanaerobium
- PMID: 39021630
- PMCID: PMC11253131
- DOI: 10.3389/fmicb.2024.1426882
Isolation and characterization of novel acetogenic strains of the genera Terrisporobacter and Acetoanaerobium
Abstract
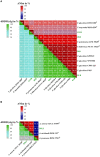
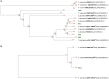
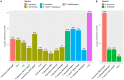
Due to their metabolic versatility in substrate utilization, acetogenic bacteria represent industrially significant production platforms for biotechnological applications such as syngas fermentation, microbial electrosynthesis or transformation of one-carbon substrates. However, acetogenic strains from the genera Terrisporobacter and Acetoanaerobium remained poorly investigated for biotechnological applications. We report the isolation and characterization of four acetogenic Terrisporobacter strains and one Acetoanaerobium strain. All Terrisporobacter isolates showed a characteristic growth pattern under a H2 + CO2 atmosphere. An initial heterotrophic growth phase was followed by a stationary growth phase, where continuous acetate production was indicative of H2-dependent acetogenesis. One of the novel Terrisporobacter isolates obtained from compost (strain COMT) additionally produced ethanol besides acetate in the stationary growth phase in H2-supplemented cultures. Genomic and physiological characterizations showed that strain COMT represented a novel Terrisporobacter species and the name Terrisporobacter vanillatitrophus is proposed (=DSM 116160T = CCOS 2104T). Phylogenomic analysis of the novel isolates and reference strains implied the reclassification of the T. petrolearius/T. hibernicus phylogenomic cluster to the species T. petrolearius and of the A. noterae/A. sticklandii phylogenomic cluster to the species A. sticklandii. Furthermore, we provide first insights into active prophages of acetogens from the genera Terrisporobacter and Acetoanaerobium.
Keywords: Acetoanaerobium; Terrisporobacter; Wood-Ljungdahl pathway; acetogen; ethanol; prophage.
Copyright © 2024 Böer, Schüler, Lüschen, Eysell, Dröge, Heinemann, Engelhardt, Basen, Daniel and Poehlein.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Genome-based metabolic and phylogenomic analysis of three Terrisporobacter species.PLoS One. 2023 Oct 10;18(10):e0290128. doi: 10.1371/journal.pone.0290128. eCollection 2023. PLoS One. 2023. PMID: 37816002 Free PMC article.
-
Isolation and characterization of novel acetogenic Moorella strains for employment as potential thermophilic biocatalysts.FEMS Microbiol Ecol. 2024 Aug 13;100(9):fiae109. doi: 10.1093/femsec/fiae109. FEMS Microbiol Ecol. 2024. PMID: 39118367 Free PMC article.
-
Formate-Dependent Acetogenic Utilization of Glucose by the Fecal Acetogen Clostridium bovifaecis.Appl Environ Microbiol. 2020 Nov 10;86(23):e01870-20. doi: 10.1128/AEM.01870-20. Print 2020 Nov 10. Appl Environ Microbiol. 2020. PMID: 32948524 Free PMC article.
-
Engineered acetogenic bacteria as microbial cell factory for diversified biochemicals.Front Bioeng Biotechnol. 2024 Jul 11;12:1395540. doi: 10.3389/fbioe.2024.1395540. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39055341 Free PMC article. Review.
-
Analysis of the Core Genome and Pan-Genome of Autotrophic Acetogenic Bacteria.Front Microbiol. 2016 Sep 28;7:1531. doi: 10.3389/fmicb.2016.01531. eCollection 2016. Front Microbiol. 2016. PMID: 27733845 Free PMC article. Review.
Cited by
-
Composite Probiotics Improve Gut Health and Enhance Tryptophan Metabolism in Nursery Piglets During Liquid Feeding.Int J Mol Sci. 2025 Jun 13;26(12):5698. doi: 10.3390/ijms26125698. Int J Mol Sci. 2025. PMID: 40565160 Free PMC article.
-
Comparative Analysis of Gut Bacteria of Four Waterbirds Species in Taolimiao-Alashan Nur (T-A Nur) in Erdos Relic Gull National Nature Reserve, Inner Mongolia, China.Ecol Evol. 2025 May 13;15(5):e71432. doi: 10.1002/ece3.71432. eCollection 2025 May. Ecol Evol. 2025. PMID: 40370353 Free PMC article.
-
High-throughput cultivation and isolation of environmental anaerobes using selectively permeable hydrogel capsules.ISME Commun. 2025 Jul 13;5(1):ycaf117. doi: 10.1093/ismeco/ycaf117. eCollection 2025 Jan. ISME Commun. 2025. PMID: 40761568 Free PMC article.
-
Exploring the Linkage Between Ruminal Microbial Communities on Postweaning and Finishing Diets and Their Relation to Residual Feed Intake in Beef Cattle.Microorganisms. 2024 Nov 27;12(12):2437. doi: 10.3390/microorganisms12122437. Microorganisms. 2024. PMID: 39770639 Free PMC article.
References
-
- Baum C., Zeldes B., Poehlein A., Daniel R., Müller V., Basen M. (2024). The energy-converting hydrogenase Ech2 is important for the growth of the thermophilic acetogen Thermoanaerobacter kivui on ferredoxin-dependent substrates. Microbiol Spectr 12:e0338023. doi: 10.1128/spectrum.03380-23, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

