Key co-expressed genes correlated with blood serum parameters of pigs fed with different fatty acid profile diets
- PMID: 39021677
- PMCID: PMC11252010
- DOI: 10.3389/fgene.2024.1394971
Key co-expressed genes correlated with blood serum parameters of pigs fed with different fatty acid profile diets
Abstract
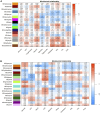
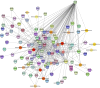
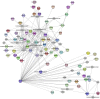
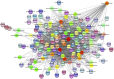
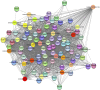
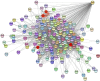
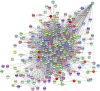
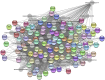
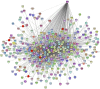
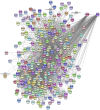
This study investigated how gene expression is affected by dietary fatty acids (FA) by using pigs as a reliable model for studying human diseases that involve lipid metabolism. This includes changes in FA composition in the liver, blood serum parameters and overall metabolic pathways. RNA-Seq data from 32 pigs were analyzed using Weighted Gene Co-expression Network Analysis (WGCNA). Our aim was to identify changes in blood serum parameters and gene expression between diets containing 3% soybean oil (SOY3.0) and a standard pig production diet containing 1.5% soybean oil (SOY1.5). Significantly, both the SOY1.5 and SOY3.0 groups showed significant modules, with a higher number of co-expressed modules identified in the SOY3.0 group. Correlated modules and specific features were identified, including enriched terms and pathways such as the histone acetyltransferase complex, type I diabetes mellitus pathway, cholesterol metabolism, and metabolic pathways in SOY1.5, and pathways related to neurodegeneration and Alzheimer's disease in SOY3.0. The variation in co-expression observed for HDL in the groups analyzed suggests different regulatory patterns in response to the higher concentration of soybean oil. Key genes co-expressed with metabolic processes indicative of diseases such as Alzheimer's was also identified, as well as genes related to lipid transport and energy metabolism, including CCL5, PNISR, DEGS1. These findings are important for understanding the genetic and metabolic responses to dietary variation and contribute to the development of more precise nutritional strategies.
Keywords: RNA-seq; WGCNA; co-expression; immune response; lipid metabolism; soybean oil; systems biology; transcriptome.
Copyright © 2024 Fanalli, Gomes, de Novais, Gervásio, Fukumasu, Moreira, Coutinho, Koltes, Amaral and Cesar.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


References
-
- Almeida V. V., Silva J. P. M., Schinckel A. P., Meira A. N., Moreira G. C. M., Gomes J. D., et al. (2021). Effects of increasing dietary oil inclusion from different sources on growth performance, carcass and meat quality traits, and fatty acid profile in genetically lean immunocastrated male pigs. Livest. Sci. 248, 104515. 10.1016/J.LIVSCI.2021.104515 - DOI
-
- Belinskaia D. A., Voronina P. A., Batalova A. A., Goncharov N. V. (2021). Serum albumin, 65–75.
LinkOut - more resources
Full Text Sources

