CMAF-Net: a cross-modal attention fusion-based deep neural network for incomplete multi-modal brain tumor segmentation
- PMID: 39022265
- PMCID: PMC11250309
- DOI: 10.21037/qims-24-9
CMAF-Net: a cross-modal attention fusion-based deep neural network for incomplete multi-modal brain tumor segmentation
Abstract
Background: The information between multimodal magnetic resonance imaging (MRI) is complementary. Combining multiple modalities for brain tumor image segmentation can improve segmentation accuracy, which has great significance for disease diagnosis and treatment. However, different degrees of missing modality data often occur in clinical practice, which may lead to serious performance degradation or even failure of brain tumor segmentation methods relying on full-modality sequences to complete the segmentation task. To solve the above problems, this study aimed to design a new deep learning network for incomplete multimodal brain tumor segmentation.
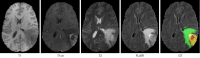
Methods: We propose a novel cross-modal attention fusion-based deep neural network (CMAF-Net) for incomplete multimodal brain tumor segmentation, which is based on a three-dimensional (3D) U-Net architecture with encoding and decoding structure, a 3D Swin block, and a cross-modal attention fusion (CMAF) block. A convolutional encoder is initially used to extract the specific features from different modalities, and an effective 3D Swin block is constructed to model the long-range dependencies to obtain richer information for brain tumor segmentation. Then, a cross-attention based CMAF module is proposed that can deal with different missing modality situations by fusing features between different modalities to learn the shared representations of the tumor regions. Finally, the fused latent representation is decoded to obtain the final segmentation result. Additionally, channel attention module (CAM) and spatial attention module (SAM) are incorporated into the network to further improve the robustness of the model; the CAM to help focus on important feature channels, and the SAM to learn the importance of different spatial regions.
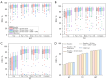
Results: Evaluation experiments on the widely-used BraTS 2018 and BraTS 2020 datasets demonstrated the effectiveness of the proposed CMAF-Net which achieved average Dice scores of 87.9%, 81.8%, and 64.3%, as well as Hausdorff distances of 4.21, 5.35, and 4.02 for whole tumor, tumor core, and enhancing tumor on the BraTS 2020 dataset, respectively, outperforming several state-of-the-art segmentation methods in missing modalities situations.
Conclusions: The experimental results show that the proposed CMAF-Net can achieve accurate brain tumor segmentation in the case of missing modalities with promising application potential.
Keywords: Brain tumor segmentation; cross-modal attention fusion (CMAF); magnetic resonance imaging (MRI); missing modalities; multimodal fusion.
2024 Quantitative Imaging in Medicine and Surgery. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-24-9/coif). The authors have no conflicts of interest to declare.
Figures








Similar articles
-
Joint learning-based feature reconstruction and enhanced network for incomplete multi-modal brain tumor segmentation.Comput Biol Med. 2023 Sep;163:107234. doi: 10.1016/j.compbiomed.2023.107234. Epub 2023 Jul 4. Comput Biol Med. 2023. PMID: 37450967
-
Multimodal Transformer of Incomplete MRI Data for Brain Tumor Segmentation.IEEE J Biomed Health Inform. 2023 Jun 16;PP. doi: 10.1109/JBHI.2023.3286689. Online ahead of print. IEEE J Biomed Health Inform. 2023. PMID: 37327094
-
Scalable Swin Transformer network for brain tumor segmentation from incomplete MRI modalities.Artif Intell Med. 2024 Mar;149:102788. doi: 10.1016/j.artmed.2024.102788. Epub 2024 Feb 2. Artif Intell Med. 2024. PMID: 38462288
-
Efficient brain tumor segmentation using Swin transformer and enhanced local self-attention.Int J Comput Assist Radiol Surg. 2024 Feb;19(2):273-281. doi: 10.1007/s11548-023-03024-8. Epub 2023 Oct 5. Int J Comput Assist Radiol Surg. 2024. PMID: 37796413 Review.
-
mResU-Net: multi-scale residual U-Net-based brain tumor segmentation from multimodal MRI.Med Biol Eng Comput. 2024 Mar;62(3):641-651. doi: 10.1007/s11517-023-02965-1. Epub 2023 Nov 19. Med Biol Eng Comput. 2024. PMID: 37981627 Review.
Cited by
-
FCFDiff-Net: full-conditional feature diffusion embedded network for 3D brain tumor segmentation.Quant Imaging Med Surg. 2025 May 1;15(5):4217-4234. doi: 10.21037/qims-24-2300. Epub 2025 Apr 25. Quant Imaging Med Surg. 2025. PMID: 40384687 Free PMC article.
References
-
- Krishna PR, Prasad V, Battula TK. Optimization empowered hierarchical residual VGGNet19 network for multi-class brain tumour classification. Multimed Tools Appl 2023;82:16691-716.
-
- Zhu Z, He X, Qi G, Li Y, Cong B, Liu Y. Brain tumor segmentation based on the fusion of deep semantics and edge information in multimodal MRI. Inf Fusion 2023;91:376-87.
LinkOut - more resources
Full Text Sources
