mRNA Nuclear Clustering Leads to a Difference in Mutant Huntingtin mRNA and Protein Silencing by siRNAs In Vivo
- PMID: 39023561
- PMCID: PMC11387003
- DOI: 10.1089/nat.2024.0027
mRNA Nuclear Clustering Leads to a Difference in Mutant Huntingtin mRNA and Protein Silencing by siRNAs In Vivo
Abstract
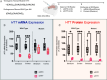
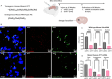
Huntington's disease (HD) is an autosomal dominant neurodegenerative disease caused by CAG repeat expansion in the first exon of the huntingtin gene (HTT). Oligonucleotide therapeutics, such as short interfering RNA (siRNA), reduce levels of huntingtin mRNA and protein in vivo and are considered a viable therapeutic strategy. However, the extent to which they silence huntingtin mRNA in the nucleus is not established. We synthesized siRNA cross-reactive to mouse (wild-type) Htt and human (mutant) HTT in a divalent scaffold and delivered to two mouse models of HD. In both models, divalent siRNA sustained lowering of wild-type Htt, but not mutant HTT mRNA expression in striatum and cortex. Near-complete silencing of both mutant HTT protein and wild-type HTT protein was observed in both models. Subsequent fluorescent in situ hybridization analysis shows that divalent siRNA acts predominantly on cytoplasmic mutant HTT transcripts, leaving clustered mutant HTT transcripts in the nucleus largely intact in treated HD mouse brains. The observed differences between mRNA and protein levels, exaggerated in the case of extended repeats, might apply to other repeat-associated neurological disorders.
Keywords: Huntington’s disease; mRNA aggregation; nuclear localization; siRNA.
Figures

Update of
-
mRNA nuclear clustering leads to a difference in mutant huntingtin mRNA and protein silencing by siRNAs in vivo.bioRxiv [Preprint]. 2024 Apr 28:2024.04.24.590997. doi: 10.1101/2024.04.24.590997. bioRxiv. 2024. Update in: Nucleic Acid Ther. 2024 Aug;34(4):164-172. doi: 10.1089/nat.2024.0027. PMID: 38774633 Free PMC article. Updated. Preprint.
References
-
- McColgan P, Tabrizi SJ. Huntington’s disease: A clinical review. Eur J Neurol 2018;25(1):24–34. - PubMed
-
- MacDonald ME, Ambrose CM, Duyao MP, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993;72(6):971–983. - PubMed
-
- Ross CA, Tabrizi SJ. Huntington’s disease: From molecular pathogenesis to clinical treatment. Lancet Neurol 2011;10(1):83–98. - PubMed
-
- Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium. Electronic address: gusella@helix.mgh.harvard.edu, Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium. CAG Repeat Not Polyglutamine Length Determines Timing of Huntington’s Disease Onset. Cell 2019;178(4):887–900.e14. - PMC - PubMed
-
- Slow EJ, van Raamsdonk J, Rogers D, et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet 2003;12(13):1555–1567. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

