Single-cell analysis reveals lasting immunological consequences of influenza infection and respiratory immunization in the pig lung
- PMID: 39024231
- PMCID: PMC11257366
- DOI: 10.1371/journal.ppat.1011910
Single-cell analysis reveals lasting immunological consequences of influenza infection and respiratory immunization in the pig lung
Abstract
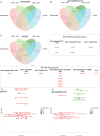
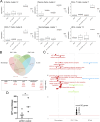
The pig is a natural host for influenza viruses and integrally involved in virus evolution through interspecies transmissions between humans and swine. Swine have many physiological, anatomical, and immunological similarities to humans, and are an excellent model for human influenza. Here, we employed single cell RNA-sequencing (scRNA-seq) and flow cytometry to characterize the major leukocyte subsets in bronchoalveolar lavage (BAL), twenty-one days after H1N1pdm09 infection or respiratory immunization with an adenoviral vector vaccine expressing hemagglutinin and nucleoprotein with or without IL-1β. Mapping scRNA-seq clusters from BAL onto those previously described in peripheral blood facilitated annotation and highlighted differences between tissue resident and circulating immune cells. ScRNA-seq data and functional assays revealed lasting impacts of immune challenge on BAL populations. First, mucosal administration of IL-1β reduced the number of functionally active Treg cells. Second, influenza infection upregulated IFI6 in BAL cells and decreased their susceptibility to virus replication in vitro. Our data provide a reference map of porcine BAL cells and reveal lasting immunological consequences of influenza infection and respiratory immunization in a highly relevant large animal model for respiratory virus infection.
Copyright: © 2024 Muir et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

