Transcriptome and microbiome-immune changes across preinvasive and invasive anal cancer lesions
- PMID: 39024554
- PMCID: PMC11343604
- DOI: 10.1172/jci.insight.180907
Transcriptome and microbiome-immune changes across preinvasive and invasive anal cancer lesions
Abstract
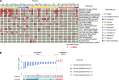
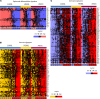
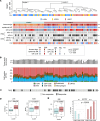
Anal squamous cell carcinoma (ASCC) is a rare gastrointestinal malignancy linked to high-risk human papillomavirus (HPV) infection, which develops from precursor lesions like low-grade squamous intraepithelial lesions and high-grade squamous intraepithelial lesions (HGSILs). ASCC incidence varies across populations and poses increased risk for people living with HIV. Our investigation focused on transcriptomic and metatranscriptomic changes from squamous intraepithelial lesions to ASCC. Metatranscriptomic analysis highlighted specific bacterial species (e.g., Fusobacterium nucleatum, Bacteroides fragilis) more prevalent in ASCC than precancerous lesions. These species correlated with gene-encoding enzymes (Acca, glyQ, eno, pgk, por) and oncoproteins (FadA, dnaK), presenting potential diagnostic or treatment markers. Unsupervised transcriptomic analysis identified distinct sample clusters reflecting histological diagnosis, immune infiltrate, HIV/HPV status, and pathway activities, recapitulating anal cancer progression's natural history. Our study unveiled molecular mechanisms in anal cancer progression, aiding in stratifying HGSIL cases based on low or high risk of progression to malignancy.
Keywords: Bioinformatics; Cell biology; Expression profiling; Molecular biology; Oncology.
Conflict of interest statement
Figures






References
-
- Darragh TM, et al. The lower anogenital squamous terminology standardization project for HPV-associated lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. Arch Pathol Lab Med. 2012;136(10):1266–1297. doi: 10.5858/arpa.LGT200570. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

