Notch signaling suppresses neuroendocrine differentiation and alters the immune microenvironment in advanced prostate cancer
- PMID: 39024561
- PMCID: PMC11364388
- DOI: 10.1172/JCI175217
Notch signaling suppresses neuroendocrine differentiation and alters the immune microenvironment in advanced prostate cancer
Abstract
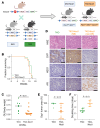
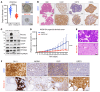
Notch signaling can have either an oncogenic or tumor-suppressive function in cancer depending on the cancer type and cellular context. While Notch can be oncogenic in early prostate cancer, we identified significant downregulation of the Notch pathway during prostate cancer progression from adenocarcinoma to neuroendocrine (NE) prostate cancer, where it functions as a tumor suppressor. Activation of Notch in NE and Rb1/Trp53-deficient prostate cancer models led to phenotypic conversion toward a more indolent, non-NE state with glandular features and expression of luminal lineage markers. This was accompanied by upregulation of MHC and type I IFN and immune cell infiltration. Overall, these data support Notch signaling as a suppressor of NE differentiation in advanced prostate cancer and provide insights into how Notch signaling influences lineage plasticity and the tumor microenvironment (TME).
Keywords: Oncology; Prostate cancer.
Figures






References
-
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

