Necroptosis enhances 'don't eat me' signal and induces macrophage extracellular traps to promote pancreatic cancer liver metastasis
- PMID: 39025845
- PMCID: PMC11258255
- DOI: 10.1038/s41467-024-50450-6
Necroptosis enhances 'don't eat me' signal and induces macrophage extracellular traps to promote pancreatic cancer liver metastasis
Abstract
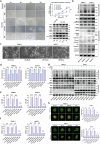
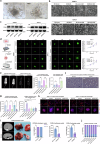
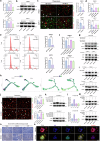
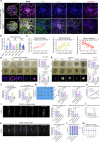
Pancreatic ductal adenocarcinoma (PDAC) is a devastating cancer with dismal prognosis due to distant metastasis, even in the early stage. Using RNA sequencing and multiplex immunofluorescence, here we find elevated expression of mixed lineage kinase domain-like pseudo-kinase (MLKL) and enhanced necroptosis pathway in PDAC from early liver metastasis T-stage (T1M1) patients comparing with non-metastatic (T1M0) patients. Mechanistically, MLKL-driven necroptosis recruits macrophages, enhances the tumor CD47 'don't eat me' signal, and induces macrophage extracellular traps (MET) formation for CXCL8 activation. CXCL8 further initiates epithelial-mesenchymal transition (EMT) and upregulates ICAM-1 expression to promote endothelial adhesion. METs also degrades extracellular matrix, that eventually supports PDAC liver metastasis. Meanwhile, targeting necroptosis and CD47 reduces liver metastasis in vivo. Our study thus reveals that necroptosis facilitates PDAC metastasis by evading immune surveillance, and also suggest that CD47 blockade, combined with MLKL inhibitor GW806742X, may be a promising neoadjuvant immunotherapy for overcoming the T1M1 dilemma and reviving the opportunity for radical surgery.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

