Timing of whole genome duplication is associated with tumor-specific MHC-II depletion in serous ovarian cancer
- PMID: 39025846
- PMCID: PMC11258338
- DOI: 10.1038/s41467-024-50137-y
Timing of whole genome duplication is associated with tumor-specific MHC-II depletion in serous ovarian cancer
Abstract
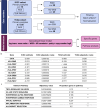
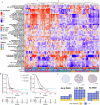
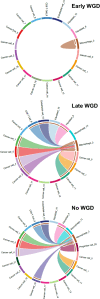
Whole genome duplication is frequently observed in cancer, and its prevalence in our prior analysis of end-stage, homologous recombination deficient high grade serous ovarian cancer (almost 80% of samples) supports the notion that whole genome duplication provides a fitness advantage under the selection pressure of therapy. Here, we therefore aim to identify potential therapeutic vulnerabilities in primary high grade serous ovarian cancer with whole genome duplication by assessing differentially expressed genes and pathways in 79 samples. We observe that MHC-II expression is lowest in tumors which have acquired whole genome duplication early in tumor evolution, and further demonstrate that reduced MHC-II expression occurs in subsets of tumor cells rather than in canonical antigen-presenting cells. Early whole genome duplication is also associated with worse patient survival outcomes. Our results suggest an association between the timing of whole genome duplication, MHC-II expression and clinical outcome in high grade serous ovarian cancer that warrants further investigation for therapeutic targeting.
© 2024. The Author(s).
Conflict of interest statement
D.D.L.B. reports research support grants from Roche-Genentech, AstraZeneca, and personal consulting fees from Exo Therapeutics, none of which are related to this work. E.L.C reports research grant funding from AstraZeneca which is not related to this work. The remaining authors declare no potential conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
- APP1161198/Department of Health | National Health and Medical Research Council (NHMRC)
- APP1189939/Department of Health | National Health and Medical Research Council (NHMRC)
- APP1161198/Department of Health | National Health and Medical Research Council (NHMRC)
- MCRF21004/Victorian Cancer Agency (VCA)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

