An intranasal nanoparticle STING agonist protects against respiratory viruses in animal models
- PMID: 39025863
- PMCID: PMC11258242
- DOI: 10.1038/s41467-024-50234-y
An intranasal nanoparticle STING agonist protects against respiratory viruses in animal models
Abstract
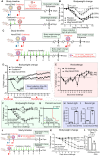
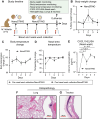
Respiratory viral infections cause morbidity and mortality worldwide. Despite the success of vaccines, vaccination efficacy is weakened by the rapid emergence of viral variants with immunoevasive properties. The development of an off-the-shelf, effective, and safe therapy against respiratory viral infections is thus desirable. Here, we develop NanoSTING, a nanoparticle formulation of the endogenous STING agonist, 2'-3' cGAMP, to function as an immune activator and demonstrate its safety in mice and rats. A single intranasal dose of NanoSTING protects against pathogenic strains of SARS-CoV-2 (alpha and delta VOC) in hamsters. In transmission experiments, NanoSTING reduces the transmission of SARS-CoV-2 Omicron VOC to naïve hamsters. NanoSTING also protects against oseltamivir-sensitive and oseltamivir-resistant strains of influenza in mice. Mechanistically, NanoSTING upregulates locoregional interferon-dependent and interferon-independent pathways in mice, hamsters, as well as non-human primates. Our results thus implicate NanoSTING as a broad-spectrum immune activator for controlling respiratory virus infection.
© 2024. The Author(s).
Conflict of interest statement
UH has filed provisional patents based on the findings of this study. N.V. and L.J.N.C. are co-founders of AuraVax Therapeutics and CellChorus. The remaining authors declare no competing interests.
Figures


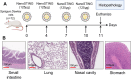

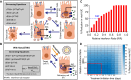

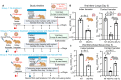
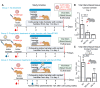
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

