A group 3 medulloblastoma stem cell program is maintained by OTX2-mediated alternative splicing
- PMID: 39025928
- PMCID: PMC11321995
- DOI: 10.1038/s41556-024-01460-5
A group 3 medulloblastoma stem cell program is maintained by OTX2-mediated alternative splicing
Abstract
OTX2 is a transcription factor and known driver in medulloblastoma (MB), where it is amplified in a subset of tumours and overexpressed in most cases of group 3 and group 4 MB. Here we demonstrate a noncanonical role for OTX2 in group 3 MB alternative splicing. OTX2 associates with the large assembly of splicing regulators complex through protein-protein interactions and regulates a stem cell splicing program. OTX2 can directly or indirectly bind RNA and this may be partially independent of its DNA regulatory functions. OTX2 controls a pro-tumorigenic splicing program that is mirrored in human cerebellar rhombic lip origins. Among the OTX2-regulated differentially spliced genes, PPHLN1 is expressed in the most primitive rhombic lip stem cells, and targeting PPHLN1 splicing reduces tumour growth and enhances survival in vivo. These findings identify OTX2-mediated alternative splicing as a major determinant of cell fate decisions that drive group 3 MB progression.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





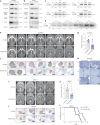


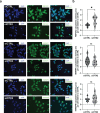

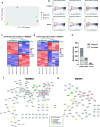


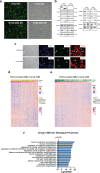
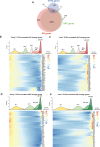
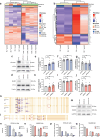
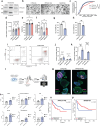
References
-
- Louis, D. N., Ohgaki, H., Wiestler, O. D. & Cavenee, W. K. WHO Classification of Tumours of the Central Nervous System 4th edn (IARC Publications, 2016).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

