Gα13 restricts nutrient driven proliferation in mucosal germinal centers
- PMID: 39025963
- PMCID: PMC11362015
- DOI: 10.1038/s41590-024-01910-0
Gα13 restricts nutrient driven proliferation in mucosal germinal centers
Abstract
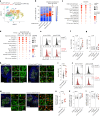
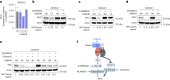
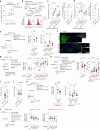
Germinal centers (GCs) that form in mucosal sites are exposed to gut-derived factors that have the potential to influence homeostasis independent of antigen receptor-driven selective processes. The G-protein Gα13 confines B cells to the GC and limits the development of GC-derived lymphoma. We discovered that Gα13-deficiency fuels the GC reaction via increased mTORC1 signaling and Myc protein expression specifically in the mesenteric lymph node (mLN). The competitive advantage of Gα13-deficient GC B cells (GCBs) in mLN was not dependent on T cell help or gut microbiota. Instead, Gα13-deficient GCBs were selectively dependent on dietary nutrients likely due to greater access to gut lymphatics. Specifically, we found that diet-derived glutamine supported proliferation and Myc expression in Gα13-deficient GCBs in the mLN. Thus, GC confinement limits the effects of dietary glutamine on GC dynamics in mucosal tissues. Gα13 pathway mutations coopt these processes to promote the gut tropism of aggressive lymphoma.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures











References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

