The actin binding protein profilin 1 localizes inside mitochondria and is critical for their function
- PMID: 39026010
- PMCID: PMC11316047
- DOI: 10.1038/s44319-024-00209-3
The actin binding protein profilin 1 localizes inside mitochondria and is critical for their function
Abstract
The monomer-binding protein profilin 1 (PFN1) plays a crucial role in actin polymerization. However, mutations in PFN1 are also linked to hereditary amyotrophic lateral sclerosis, resulting in a broad range of cellular pathologies which cannot be explained by its primary function as a cytosolic actin assembly factor. This implies that there are important, undiscovered roles for PFN1 in cellular physiology. Here we screened knockout cells for novel phenotypes associated with PFN1 loss of function and discovered that mitophagy was significantly upregulated. Indeed, despite successful autophagosome formation, fusion with the lysosome, and activation of additional mitochondrial quality control pathways, PFN1 knockout cells accumulate depolarized, dysmorphic mitochondria with altered metabolic properties. Surprisingly, we also discovered that PFN1 is present inside mitochondria and provide evidence that mitochondrial defects associated with PFN1 loss are not caused by reduced actin polymerization in the cytosol. These findings suggest a previously unrecognized role for PFN1 in maintaining mitochondrial integrity and highlight new pathogenic mechanisms that can result from PFN1 dysregulation.
Keywords: Actin; Mitochondria; Mitochondrial-derived Vesicles; Mitophagy; Profilin.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
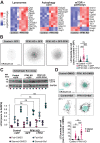

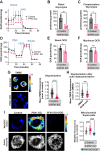
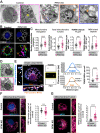


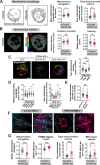

Update of
-
The actin binding protein profilin 1 is critical for mitochondria function.bioRxiv [Preprint]. 2023 Sep 19:2023.08.07.552354. doi: 10.1101/2023.08.07.552354. bioRxiv. 2023. Update in: EMBO Rep. 2024 Aug;25(8):3240-3262. doi: 10.1038/s44319-024-00209-3. PMID: 37609280 Free PMC article. Updated. Preprint.
References
-
- Abd Elghani F, Safory H, Hamza H, Savyon M, Farhoud M, Toren-Hershoviz M, Vitic Z, Ebanks K, Shani V, Bisharat S et al (2022) SIAH proteins regulate the degradation and intra-mitochondrial aggregation of PINK1: implications for mitochondrial pathology in Parkinson’s disease. Aging Cell 21:e13731 10.1111/acel.13731 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

