Spatial tumor immune microenvironment phenotypes in ovarian cancer
- PMID: 39026018
- PMCID: PMC11258306
- DOI: 10.1038/s41698-024-00640-8
Spatial tumor immune microenvironment phenotypes in ovarian cancer
Abstract
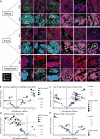
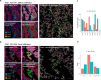
Immunotherapy has largely failed in ovarian carcinoma (OC), likely due to that the vast tumor heterogeneity and variation in immune response have hampered clinical trial outcomes. Tumor-immune microenvironment (TIME) profiling may aid in stratification of OC tumors for guiding treatment selection. Here, we used Digital Spatial Profiling combined with image analysis to characterize regions of spatially distinct TIME phenotypes in OC to assess whether immune infiltration pattern can predict presence of immuno-oncology targets. Tumors with diffuse immune infiltration and increased tumor-immune spatial interactions had higher presence of IDO1, PD-L1, PD-1 and Tim-3, while focal immune niches had more CD163 macrophages and a preliminary worse outcome. Immune exclusion was associated with presence of Tregs and Fibronectin. High-grade serous OC showed an overall stronger immune response and presence of multiple targetable checkpoints. Low-grade serous OC was associated with diffuse infiltration and a high expression of STING, while endometrioid OC had higher presence of CTLA-4. Mucinous and clear cell OC were dominated by focal immune clusters and immune-excluded regions, with mucinous tumors displaying T-cell rich immune niches.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Banerjee S, et al. Maintenance olaparib for patients with newly diagnosed advanced ovarian cancer and a BRCA mutation (SOLO1/GOG 3004): 5-year follow-up of a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2021;22:1721–1731. doi: 10.1016/S1470-2045(21)00531-3. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

