Adipose-derived stem cell exosomes loaded with icariin alleviates rheumatoid arthritis by modulating macrophage polarization in rats
- PMID: 39026367
- PMCID: PMC11256651
- DOI: 10.1186/s12951-024-02711-1
Adipose-derived stem cell exosomes loaded with icariin alleviates rheumatoid arthritis by modulating macrophage polarization in rats
Abstract
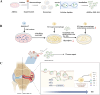
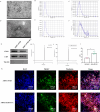
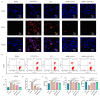
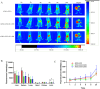
Rheumatoid arthritis (RA) is a chronic autoimmune disease marked by synovitis and cartilage destruction. The active compound, icariin (ICA), derived from the herb Epimedium, exhibits potent anti-inflammatory properties. However, its clinical utility is limited by its water insolubility, poor permeability, and low bioavailability. To address these challenges, we developed a multifunctional drug delivery system-adipose-derived stem cells-exosomes (ADSCs-EXO)-ICA to target active macrophages in synovial tissue and modulate macrophage polarization from M1 to M2. High-performance liquid chromatography analysis confirmed a 92.4 ± 0.008% loading efficiency for ADSCs-EXO-ICA. In vitro studies utilizing cellular immunofluorescence (IF) and flow cytometry demonstrated significant inhibition of M1 macrophage proliferation by ADSCs-EXO-ICA. Enzyme-linked immunosorbent assay, cellular transcriptomics, and real-time quantitative PCR indicated that ADSCs-EXO-ICA promotes an M1-to-M2 phenotypic transition by reducing glycolysis through the inhibition of the ERK/HIF-1α/GLUT1 pathway. In vivo, ADSCs-EXO-ICA effectively accumulated in the joints. Pharmacodynamic assessments revealed that ADSCs-EXO-ICA decreased cytokine levels and mitigated arthritis symptoms in collagen-induced arthritis (CIA) rats. Histological analysis and micro computed tomography confirmed that ADSCs-EXO-ICA markedly ameliorated synovitis and preserved cartilage. Further in vivo studies indicated that ADSCs-EXO-ICA suppresses arthritis by promoting an M1-to-M2 switch and suppressing glycolysis. Western blotting supported the therapeutic efficacy of ADSCs-EXO-ICA in RA, confirming its role in modulating macrophage function through energy metabolism regulation. Thus, this study not only introduces a drug delivery system that significantly enhances the anti-RA efficacy of ADSCs-EXO-ICA but also elucidates its mechanism of action in macrophage function inhibition.
Keywords: A novel drug delivery system—adipose-derived stem cells-exosomes (ADSCs-EXO)-ICA; Adipose-derived stem cell exosomes; Collagen-induced arthritis; Macrophage polarization; Rheumatoid arthritis; Synergistic effect; icariin.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Boutet MA, Courties G, Nerviani A, Le Goff B, Apparailly F, Pitzalis C. F. Blanchard Autoimmun Rev 2021,20,102758. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

