Synthesis, biological evaluation and metadynamics simulations of novel N-methyl β-sheet breaker peptides as inhibitors of Alzheimer's β-amyloid fibrillogenesis
- PMID: 39026638
- PMCID: PMC11253850
- DOI: 10.1039/d4md00057a
Synthesis, biological evaluation and metadynamics simulations of novel N-methyl β-sheet breaker peptides as inhibitors of Alzheimer's β-amyloid fibrillogenesis
Abstract
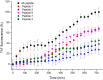
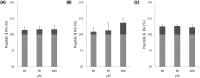
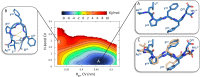
Several scientific evidences report that a central role in the pathogenesis of Alzheimer's disease is played by the deposition of insoluble aggregates of β-amyloid proteins in the brain. Because Aβ is self-assembling, one possible design strategy is to inhibit the aggregation of Aβ peptides using short peptide fragments homologous to the full-length wild-type Aβ protein. In the past years, several studies have reported on the synthesis of some short synthetic peptides called β-sheet breaker peptides (BSBPs). Herein, we present the synthesis of novel (cell-permeable) N-methyl BSBPs, designed based on literature information on the structural key features of BSBPs. Three-dimensional GRID-based pharmacophore peptide screening combined with PT-WTE metadynamics was performed to support the results of the design and microwave-assisted synthesis of peptides 2 and 3 prepared and analyzed for their fibrillogenesis inhibition activity and cytotoxicity. An HR-MS-based cell metabolomic approach highlighted their cell permeability properties.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures





References
LinkOut - more resources
Full Text Sources
Miscellaneous

