This is a preprint.
Antagonistic Roles for MITF and TFE3 in Melanoma Plasticity
- PMID: 39026725
- PMCID: PMC11257520
- DOI: 10.1101/2024.07.11.603140
Antagonistic Roles for MITF and TFE3 in Melanoma Plasticity
Update in
-
Antagonistic roles for MITF and TFE3 in melanoma plasticity.Cell Rep. 2025 Apr 22;44(4):115474. doi: 10.1016/j.celrep.2025.115474. Epub 2025 Mar 25. Cell Rep. 2025. PMID: 40138313 Free PMC article.
Abstract
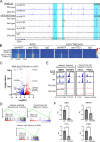
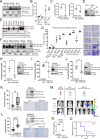
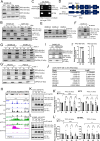
Melanoma cells have the ability to switch from a melanocytic and proliferative state to a mesenchymal and invasive state and back again. This plasticity drives intra-tumoral heterogeneity, progression, and therapeutic resistance. Microphthalmia-associated Transcription Factor (MITF) promotes the melanocytic/proliferative phenotype, but factors that drive the mesenchymal/invasive phenotype and the mechanisms that effect the switch between cell states are unclear. Here, we identify the MITF paralog TFE3 and the non-canonical mTORC1 pathway as regulators of the mesenchymal state. We show that TFE3 expression drives the metastatic phenotype in melanoma cell lines and tumors. Deletion of TFE3 in MITF-low melanoma cell lines suppresses their ability to migrate and metastasize. Further, MITF suppresses the mesenchymal phenotype by directly or indirectly activating expression of FNIP1, FNIP2, and FLCN, which encode components of the non-canonical mTORC1 pathway, thereby promoting cytoplasmic retention and lysosome-mediated degradation of TFE3. These findings highlight a molecular pathway controlling melanoma plasticity and invasiveness.
Keywords: MITF; Melanoma; TFE3; cell plasticity; mTORC1; metastasis; phenotype-switching; protein stability.
Conflict of interest statement
Declaration of Interests: All authors declare no competing interests.
Figures


References
-
- Wouters J., Kalender-Atak Z., Minnoye L., Spanier K.I., De Waegeneer M., Bravo González-Blas C., Mauduit D., Davie K., Hulselmans G., Najem A., et al. (2020). Robust gene expression programs underlie recurrent cell states and phenotype switching in melanoma. Nat Cell Biol 22, 986–998. 10.1038/s41556-020-0547-3. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
