This is a preprint.
Efficient Genome Editing with Chimeric Oligonucleotide-Directed Editing
- PMID: 39026836
- PMCID: PMC11257564
- DOI: 10.1101/2024.07.09.602710
Efficient Genome Editing with Chimeric Oligonucleotide-Directed Editing
Abstract
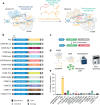
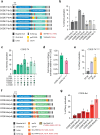
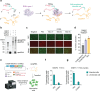
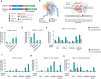
Prime editing has emerged as a precise and powerful genome editing tool, offering a favorable gene editing profile compared to other Cas9-based approaches. Here we report new nCas9-DNA polymerase fusion proteins to create chimeric oligonucleotide-directed editing (CODE) systems for search-and-replace genome editing. Through successive rounds of engineering, we developed CODEMax and CODEMax(exo+) editors that achieve efficient genome modifications in human cells with low unintended edits. CODEMax and CODEMax(exo+) contain an engineered Bst DNA polymerase derivative known for its robust strand displacement ability. Additionally, CODEMax(exo+) features a 5' to 3' exonuclease activity that promotes effective strand invasion and repair outcomes favoring the incorporation of the desired edit. We demonstrate CODEs can perform small insertions, deletions, and substitutions with improved efficiency compared to PEMax at many loci. Overall, CODEs complement existing prime editors to expand the toolbox for genome manipulations without double-stranded breaks.
Conflict of interest statement
L.N., N.R., B.P., C.M., and P.K.J. are listed as inventors on the patent applications related to the content of this work. P.K.J. is a co-founder of CasNx, LLC, Par Biosciences, LLC, and CRISPR, LLC. C.M. is a co-founder of Carver Biosciences. B.A. is an advisory board member with options for Arbor Biotechnologies and Tessera Therapeutics. B.A. holds equity in Celsius Therapeutics. J.Y. and B.A. have filed patent application(s) related to prime editing and/or other CRISPR-based technologies. J.E.T. is a scientific advisor for Prolific Machines and Nereid Therapeutics.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
