This is a preprint.
Heli-SMACC: Helicase-targeting SMAll Molecule Compound Collection
- PMID: 39026851
- PMCID: PMC11257486
- DOI: 10.1101/2024.07.04.602122
Heli-SMACC: Helicase-targeting SMAll Molecule Compound Collection
Abstract
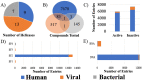
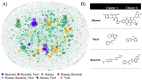
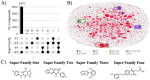
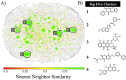
Helicases have emerged as promising targets for the development of antiviral drugs; however, the family remains largely undrugged. To support the focused development of viral helicase inhibitors we identified, collected, and integrated all chemogenomics data for all available helicases from the ChEMBL database. After thoroughly curating and enriching the data with relevant annotations we have created a derivative database of helicase inhibitors which we dubbed Heli-SMACC (Helicase-targeting SMAll Molecule Compound Collection). The current version of Heli-SMACC contains 20,432 bioactivity entries for viral, human, and bacterial helicases. We have selected 30 compounds with promising viral helicase activity and tested them in a SARS-CoV-2 NSP13 ATPase assay. Twelve compounds demonstrated ATPase inhibition and a consistent dose-response curve. The Heli-SMACC database may serve as a reference for virologists and medicinal chemists working on the development of novel helicase inhibitors. Heli-SMACC is publicly available at https://smacc.mml.unc.edu.
Keywords: Antiviral Drug Discovery; Database; Drug Repurposing; Helicases; Viral Helicase Inhibitors.
Conflict of interest statement
Conflict of interest AT and ENM are co-founders of Predictive, LLC, which develops novel alternative methods and software for toxicity prediction. All other authors have nothing to disclose.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
