This is a preprint.
Cerebellar output neurons impair non-motor behaviors by altering development of extracerebellar connectivity
- PMID: 39026865
- PMCID: PMC11257463
- DOI: 10.1101/2024.07.08.602496
Cerebellar output neurons impair non-motor behaviors by altering development of extracerebellar connectivity
Update in
-
Cerebellar output neurons can impair non-motor behaviors by altering development of extracerebellar connectivity.Nat Commun. 2025 Feb 21;16(1):1858. doi: 10.1038/s41467-025-57080-6. Nat Commun. 2025. PMID: 39984491 Free PMC article.
Abstract
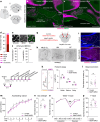
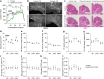
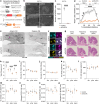
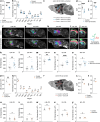
The capacity of the brain to compensate for insults during development depends on the type of cell loss, whereas the consequences of genetic mutations in the same neurons are difficult to predict. We reveal powerful compensation from outside the cerebellum when the excitatory cerebellar output neurons are ablated embryonically and demonstrate that the minimum requirement for these neurons is for motor coordination and not learning and social behaviors. In contrast, loss of the homeobox transcription factors Engrailed1/2 (EN1/2) in the cerebellar excitatory lineage leads to additional deficits in adult learning and spatial working memory, despite half of the excitatory output neurons being intact. Diffusion MRI indicates increased thalamo-cortico-striatal connectivity in En1/2 mutants, showing that the remaining excitatory neurons lacking En1/2 exert adverse effects on extracerebellar circuits regulating motor learning and select non-motor behaviors. Thus, an absence of cerebellar output neurons is less disruptive than having cerebellar genetic mutations.
Conflict of interest statement
Competing interests All authors declare to have no actual or potential conflict of interest including any financial, personal, or other relationships with other people or organizations within three years of beginning the submitted work that could inappropriately influence, or be perceived to influence, their work.
Figures




References
-
- Korah MP, et al. Incidence, risks, and sequelae of posterior fossa syndrome in pediatric medulloblastoma. Int J Radiat Oncol Biol Phys 77, 106–112 (2010). - PubMed
-
- Robertson PL, et al. Incidence and severity of postoperative cerebellar mutism syndrome in children with medulloblastoma: a prospective study by the Children’s Oncology Group. J Neurosurg 105, 444–451 (2006). - PubMed
Publication types
Grants and funding
- R01 DA050454/DA/NIDA NIH HHS/United States
- R01 MH112143/MH/NIMH NIH HHS/United States
- TL1 TR002386/TR/NCATS NIH HHS/United States
- R01 DA053261/DA/NIDA NIH HHS/United States
- R01 MH110553/MH/NIMH NIH HHS/United States
- R01 MH125006/MH/NIMH NIH HHS/United States
- R37 MH085726/MH/NIMH NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 NS102904/NS/NINDS NIH HHS/United States
- T32 HD060600/HD/NICHD NIH HHS/United States
- R01 NS116137/NS/NINDS NIH HHS/United States
- R01 MH118934/MH/NIMH NIH HHS/United States
- R01 DA029122/DA/NIDA NIH HHS/United States
- R01 DA054368/DA/NIDA NIH HHS/United States
- R01 NS092096/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
