Accounting for cellular-level variation in lysis: implications for virus-host dynamics
- PMID: 39028198
- PMCID: PMC11323501
- DOI: 10.1128/mbio.01376-24
Accounting for cellular-level variation in lysis: implications for virus-host dynamics
Abstract
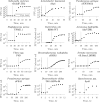
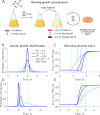
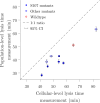
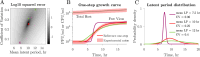
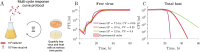
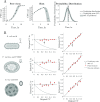
Viral impacts on microbial populations depend on interaction phenotypes-including viral traits spanning the adsorption rate, latent period, and burst size. The latent period is a key viral trait in lytic infections. Defined as the time from viral adsorption to viral progeny release, the latent period of bacteriophage is conventionally inferred via one-step growth curves in which the accumulation of free virus is measured over time in a population of infected cells. Developed more than 80 years ago, one-step growth curves do not account for cellular-level variability in the timing of lysis, potentially biasing inference of viral traits. Here, we use nonlinear dynamical models to understand how individual-level variation of the latent period impacts virus-host dynamics. Our modeling approach shows that inference of the latent period via one-step growth curves is systematically biased-generating estimates of shorter latent periods than the underlying population-level mean. The bias arises because variability in lysis timing at the cellular level leads to a fraction of early burst events, which are interpreted, artefactually, as an earlier mean time of viral release. We develop a computational framework to estimate latent period variability from joint measurements of host and free virus populations. Our computational framework recovers both the mean and variance of the latent period within simulated infections including realistic measurement noise. This work suggests that reframing the latent period as a distribution to account for variability in the population will improve the study of viral traits and their role in shaping microbial populations.IMPORTANCEQuantifying viral traits-including the adsorption rate, burst size, and latent period-is critical to characterize viral infection dynamics and develop predictive models of viral impacts across scales from cells to ecosystems. Here, we revisit the gold standard of viral trait estimation-the one-step growth curve-to assess the extent to which assumptions at the core of viral infection dynamics lead to ongoing and systematic biases in inferences of viral traits. We show that latent period estimates obtained via one-step growth curves systematically underestimate the mean latent period and, in turn, overestimate the rate of viral killing at population scales. By explicitly incorporating trait variability into a dynamical inference framework that leverages both virus and host time series, we provide a practical route to improve estimates of the mean and variance of viral traits across diverse virus-microbe systems.
Keywords: bacteriophage lysis; cellular variability; inference; latent period; mathematical modeling; phage ecology; population dynamics; viral traits.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Levin BR, Stewart FM, Chao L. 1977. Resource-limited growth, competition, and predation: a model and experimental studies with bacteria and bacteriophage. Amer Nat 111:3–24. doi:10.1086/283134 - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
