Trophoblast Side-Population Markers are Dysregulated in Preeclampsia and Fetal Growth Restriction
- PMID: 39028417
- PMCID: PMC11445292
- DOI: 10.1007/s12015-024-10764-w
Trophoblast Side-Population Markers are Dysregulated in Preeclampsia and Fetal Growth Restriction
Abstract
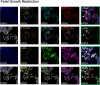
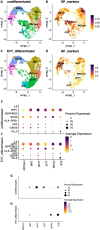
Dysregulated progenitor cell populations may contribute to poor placental development and placental insufficiency pathogenesis. Side-population cells possess progenitor properties. Recent human trophoblast side-population isolation identified enrichment of 8 specific genes (CXCL8, ELL2, GATA6, HK2, HLA-DPB1, INTS6, SERPINE3 and UPP1) (Gamage et al. 2020, Stem Cell Rev Rep). We characterised these trophoblast side-population markers in human placenta and in placental insufficiency disorders: preeclampsia and fetal growth restriction (FGR). Trophoblast side-population markers localised to mononuclear trophoblasts lining the placental villous basement membrane in preterm control, preeclamptic and FGR placental sections (n = 3, panel of 3 markers/serial section). Analysis of single-cell transcriptomics of an organoid human trophoblast stem cell (hTSC) to extravillous trophoblast (EVT) differentiation model (Shannon et al. 2022, Development) identified that all side-population genes were enriched in mononuclear trophoblast and trophoblasts committed to differentiation under hTSC culture conditions. In vitro validation via 96 h time course hTSC differentiation to EVTs or syncytiotrophoblasts (n = 5) demonstrated ELL2 and HK2 increased with differentiation (p < 0.0024, p < 0.0039 respectively). CXCL8 and HLA-DPB1 were downregulated (p < 0.030, p < 0.011 respectively). GATA6 and INTS6 increased with EVT differentiation only, and UPP1 reduced with syncytialisation. SERPINE3 was undetectable. Trophoblast side-population marker mRNA was measured in human placentas (< 34-weeks' gestation; n = 78 preeclampsia, n = 30 FGR, and n = 18 gestation-matched controls). ELL2, HK2 and CXCL8 were elevated in preeclamptic (p = 0.0006, p < 0.0001, p = 0.0335 respectively) and FGR placentas (p = 0.0065, p < 0.0001, p = 0.0001 respectively) versus controls. Placental GATA6 was reduced in pregnancies with preeclampsia and FGR (p = 0.0014, p = 0.0146 respectively). Placental INTS6 was reduced with FGR only (p < 0.0001). This study identified the localisation of a unique trophoblast subset enriched for side-population markers. Aberrant expression of some side-population markers may indicate disruptions to unique trophoblast subtypes in placental insufficiency.
Keywords: Fetal growth restriction; Placenta; Preeclampsia; Pregnancy; Trophoblast.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest nor competing interests.
Figures




References
-
- Abalos, E., Cuesta, C., Grosso, A. L., Chou, D., & Say, L. (2013). Global and regional estimates of preeclampsia and eclampsia: A systematic review. European Journal of Obstetrics & Gynecology and Reproductive Biology,170(1), 1–7. - PubMed
-
- Dimitriadis, E., Rolnik, D. L., Zhou, W., Estrada-Gutierrez, G., Koga, K., Francisco, R. P. V., Whitehead, C., Hyett, J., da Silva, C. F., Nicolaides, K., & Menkhorst, E. (2023). Pre-eclampsia. Nature Reviews Disease Primers,9(1), 8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

