Small-molecule probe for IBD risk variant GPR65 I231L alters cytokine signaling networks through positive allosteric modulation
- PMID: 39028811
- PMCID: PMC11259170
- DOI: 10.1126/sciadv.adn2339
Small-molecule probe for IBD risk variant GPR65 I231L alters cytokine signaling networks through positive allosteric modulation
Abstract
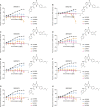
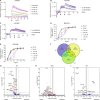
The proton-sensing heterotrimeric guanine nucleotide-binding protein-coupled receptor GPR65 is expressed in immune cells and regulates tissue homeostasis in response to decreased extracellular pH, which occurs in the context of inflammation and tumorigenesis. Genome-wide association studies linked GPR65 to several autoimmune and inflammatory diseases such as multiple sclerosis and inflammatory bowel disease (IBD). The loss-of-function GPR65 I231L IBD risk variant alters cellular metabolism, impairs protective tissue functions, and increases proinflammatory cytokine production. Hypothesizing that a small molecule designed to potentiate GPR65 at subphysiological pH could decrease inflammatory responses, we found positive allosteric modulators of GPR65 that engage and activate both human and mouse orthologs of the receptor. We observed that the chemical probe BRD5075 alters cytokine and chemokine programs in dendritic cells, establishing that immune signaling can be modulated by targeting GPR65. Our investigation offers improved chemical probes to further interrogate the biology of human GPR65 and its clinically relevant genetic variants.
Figures


References
-
- Nelson M. R., Tipney H., Painter J. L., Shen J., Nicoletti P., Shen Y., Floratos A., Sham P. C., Li M. J., Wang J., Cardon L. R., Whittaker J. C., Sanseau P., The support of human genetic evidence for approved drug indications. Nat. Genet. 47, 856–860 (2015). - PubMed
-
- Kong L., Pokatayev V., Lefkovith A., Carter G. T., Creasey E. A., Krishna C., Subramanian S., Kochar B., Ashenberg O., Lau H., Ananthakrishnan A. N., Graham D. B., Deguine J., Xavier R. J., The landscape of immune dysregulation in Crohn’s disease revealed through single-cell transcriptomic profiling in the ileum and colon. Immunity 56, 444–458.e5 (2023). - PMC - PubMed
-
- Smillie C. S., Biton M., Ordovas-Montanes J., Sullivan K. M., Burgin G., Graham D. B., Herbst R. H., Rogel N., Slyper M., Waldman J., Sud M., Andrews E., Velonias G., Haber A. L., Jagadeesh K., Vickovic S., Yao J., Stevens C., Dionne D., Nguyen L. T., Villani A.-C., Hofree M., Creasey E. A., Huang H., Rozenblatt-Rosen O., Garber J. J., Khalili H., Desch A. N., Daly M. J., Ananthakrishnan A. N., Shalek A. K., Xavier R. J., Regev A., Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730.e22 (2019). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

