R-loop resolution by ARIP4 helicase promotes androgen-mediated transcription induction
- PMID: 39028815
- PMCID: PMC11259169
- DOI: 10.1126/sciadv.adm9577
R-loop resolution by ARIP4 helicase promotes androgen-mediated transcription induction
Abstract
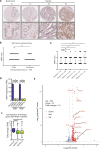
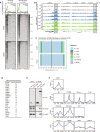
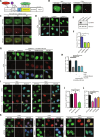
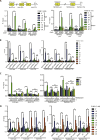
Pausing of RNA polymerase II (Pol II) at transcription start sites (TSSs) primes target genes for productive elongation. Coincidentally, DNA double-strand breaks (DSBs) enrich at highly transcribed and Pol II-paused genes, although their interplay remains undefined. Using androgen receptor (AR) signaling as a model, we have uncovered AR-interacting protein 4 (ARIP4) helicase as a driver of androgen-dependent transcription induction. Chromatin immunoprecipitation sequencing analysis revealed that ARIP4 preferentially co-occupies TSSs with paused Pol II. Moreover, we found that ARIP4 complexes with topoisomerase II beta and mediates transient DSB formation upon hormone stimulation. Accordingly, ARIP4 deficiency compromised release of paused Pol II and resulted in R-loop accumulation at a panel of highly transcribed AR target genes. Last, we showed that ARIP4 binds and unwinds R-loops in vitro and that its expression positively correlates with prostate cancer progression. We propose that androgen stimulation triggers ARIP4-mediated unwinding of R-loops at TSSs, enforcing Pol II pause release to effectively drive an androgen-dependent expression program.
Figures


References
-
- Huggins C., Hodges C. V., Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J. Clin. 22, 232–240 (1972). - PubMed
-
- Tomlins S. A., Rhodes D. R., Perner S., Dhanasekaran S. M., Mehra R., Sun X. W., Varambally S., Cao X., Tchinda J., Kuefer R., Lee C., Montie J. E., Shah R. B., Pienta K. J., Rubin M. A., Chinnaiyan A. M., Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

