Haploinsufficiency in PTPN2 leads to early-onset systemic autoimmunity from Evans syndrome to lupus
- PMID: 39028869
- PMCID: PMC11259789
- DOI: 10.1084/jem.20232337
Haploinsufficiency in PTPN2 leads to early-onset systemic autoimmunity from Evans syndrome to lupus
Abstract
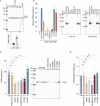
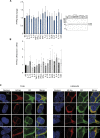
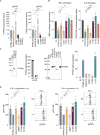
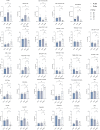
An exome sequencing strategy employed to identify pathogenic variants in patients with pediatric-onset systemic lupus or Evans syndrome resulted in the discovery of six novel monoallelic mutations in PTPN2. PTPN2 is a phosphatase that acts as an essential negative regulator of the JAK/STAT pathways. All mutations led to a loss of PTPN2 regulatory function as evidenced by in vitro assays and by hyperproliferation of patients' T cells. Furthermore, patients exhibited high serum levels of inflammatory cytokines, mimicking the profile observed in individuals with gain-of-function mutations in STAT factors. Flow cytometry analysis of patients' blood cells revealed typical alterations associated with autoimmunity and all patients presented with autoantibodies. These findings further supported the notion that a loss of function in negative regulators of cytokine pathways can lead to a broad spectrum of autoimmune manifestations and that PTPN2 along with SOCS1 haploinsufficiency constitute a new group of monogenic autoimmune diseases that can benefit from targeted therapy.
© 2024 Jeanpierre et al.
Conflict of interest statement
Disclosures: E. Crickx reported personal fees from Novartis, Amgen, UCB, and Sanofi outside the submitted work. A. Belot reported grants from Boehringer Ingelheim and personal fees from Abbvie, Kabi, and GlaxoSmithKline during the conduct of the study. No other disclosures were reported.
Figures






Comment in
-
PTPN2 deficiency: Amping up JAK/STAT.J Exp Med. 2024 Sep 2;221(9):e20240980. doi: 10.1084/jem.20240980. Epub 2024 Jul 19. J Exp Med. 2024. PMID: 39028870 Free PMC article.
References
-
- Baumgartner, C.K., Ebrahimi-Nik H., Iracheta-Vellve A., Hamel K.M., Olander K.E., Davis T.G.R., McGuire K.A., Halvorsen G.T., Avila O.I., Patel C.H., et al. . 2023. The PTPN2/PTPN1 inhibitor ABBV-CLS-484 unleashes potent anti-tumour immunity. Nature. 622:850–862. 10.1038/s41586-023-06575-7 - DOI - PMC - PubMed
-
- Del Bel, K.L., Ragotte R.J., Saferali A., Lee S., Vercauteren S.M., Mostafavi S.A., Schreiber R.A., Prendiville J.S., Phang M.S., Halparin J., et al. . 2017. JAK1 gain-of-function causes an autosomal dominant immune dysregulatory and hypereosinophilic syndrome. J. Allergy Clin. Immunol. 139:2016–2020.e5. 10.1016/j.jaci.2016.12.957 - DOI - PubMed
-
- Denoth, L., Juillerat P., Kremer A.E., Rogler G., Scharl M., Yilmaz B., Bluemel S., and On Behalf Of The Swiss Ibd Cohort Study . 2021. Modulation of the mucosa-associated microbiome linked to the PTPN2 risk gene in patients with primary sclerosing cholangitis and ulcerative colitis. Microorganisms. 9:1752. 10.3390/microorganisms9081752 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

