Dynamics in Cre-loxP site-specific recombination
- PMID: 39029281
- PMCID: PMC11616326
- DOI: 10.1016/j.sbi.2024.102878
Dynamics in Cre-loxP site-specific recombination
Abstract
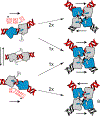
Cre recombinase is a phage-derived enzyme that has found utility for precise manipulation of DNA sequences. Cre recognizes and recombines pairs of loxP sequences characterized by an inverted repeat and asymmetric spacer. Cre cleaves and religates its DNA targets such that error-prone repair pathways are not required to generate intact DNA products. Major obstacles to broader applications are lack of knowledge of how Cre recognizes its targets, and how its activity is controlled. The picture emerging from high resolution methods is that the dynamic properties of both the enzyme and its DNA target are important determinants of its activity in both sequence recognition and DNA cleavage. Improved understanding of the role of dynamics in the key steps along the pathway of Cre-loxP recombination should significantly advance our ability to both redirect Cre to new sequences and to control its DNA cleavage activity in the test tube and in cells.
Copyright © 2024 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


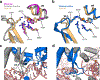

Similar articles
-
Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation.Nucleic Acids Res. 2003 Sep 15;31(18):5449-60. doi: 10.1093/nar/gkg732. Nucleic Acids Res. 2003. PMID: 12954782 Free PMC article.
-
Engineering spacer specificity of the Cre/loxP system.Nucleic Acids Res. 2024 Jul 22;52(13):8017-8031. doi: 10.1093/nar/gkae481. Nucleic Acids Res. 2024. PMID: 38869070 Free PMC article.
-
Principles of site-specific recombinase (SSR) technology.J Vis Exp. 2008 May 29;(15):718. doi: 10.3791/718. J Vis Exp. 2008. PMID: 19066587 Free PMC article.
-
Cre Recombinase.Microbiol Spectr. 2015 Feb;3(1):MDNA3-0014-2014. doi: 10.1128/microbiolspec.MDNA3-0014-2014. Microbiol Spectr. 2015. PMID: 26104563 Review.
-
Cre-loxP biochemistry.Methods. 2002 Nov;28(3):374-83. doi: 10.1016/s1046-2023(02)00244-x. Methods. 2002. PMID: 12431441 Review.
Cited by
-
Protein and DNA Conformational Changes Contribute to Specificity of Cre Recombinase.bioRxiv [Preprint]. 2024 Dec 11:2024.12.11.627928. doi: 10.1101/2024.12.11.627928. bioRxiv. 2024. Update in: Biochemistry. 2025 Mar 04;64(5):1055-1064. doi: 10.1021/acs.biochem.4c00841. PMID: 39713331 Free PMC article. Updated. Preprint.
-
Rapid in vitro method to assemble and transfer DNA fragments into the JCVI-syn3B minimal synthetic bacterial genome through Cre/loxP system.Biophys Physicobiol. 2024 Nov 7;21(4):e210024. doi: 10.2142/biophysico.bppb-v21.0024. eCollection 2024. Biophys Physicobiol. 2024. PMID: 39963596 Free PMC article.
References
-
- Craig NL: The Mechanism of Conservative Site-Specific Recombination. Annu Rev Genet 1988, 22:77–105. - PubMed
-
- Grindley NDF, Whiteson KL, Rice PA: Mechanisms of site-specific recombination. Annu Rev Biochem 2006, 75:567–605. - PubMed
-
- Meinke G, Bohm A, Hauber J, Pisabarro MT, Buchholz F: Cre Recombinase and Other Tyrosine Recombinases. Chem Rev 2016, 116:12785–12820. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

