Pan-serotype dengue virus inhibitor JNJ-A07 targets NS4A-2K-NS4B interaction with NS2B/NS3 and blocks replication organelle formation
- PMID: 39030239
- PMCID: PMC11271582
- DOI: 10.1038/s41467-024-50437-3
Pan-serotype dengue virus inhibitor JNJ-A07 targets NS4A-2K-NS4B interaction with NS2B/NS3 and blocks replication organelle formation
Abstract
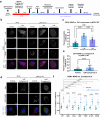
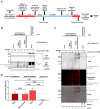
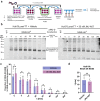
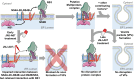
Dengue fever represents a significant medical and socio-economic burden in (sub)tropical regions, yet antivirals for treatment or prophylaxis are lacking. JNJ-A07 was described as highly active against the different genotypes within each serotype of the disease-causing dengue virus (DENV). Based on clustering of resistance mutations it has been assumed to target DENV non-structural protein 4B (NS4B). Using a photoaffinity labeling compound with high structural similarity to JNJ-A07, here we demonstrate binding to NS4B and its precursor NS4A-2K-NS4B. Consistently, we report recruitment of the compound to intracellular sites enriched for these proteins. We further specify the mechanism-of-action of JNJ-A07, which has virtually no effect on viral polyprotein cleavage, but targets the interaction between the NS2B/NS3 protease/helicase complex and the NS4A-2K-NS4B cleavage intermediate. This interaction is functionally linked to de novo formation of vesicle packets (VPs), the sites of DENV RNA replication. JNJ-A07 blocks VPs biogenesis with little effect on established ones. A similar mechanism-of-action was found for another NS4B inhibitor, NITD-688. In summary, we unravel the antiviral mechanism of these NS4B-targeting molecules and show how DENV employs a short-lived cleavage intermediate to carry out an early step of the viral life cycle.
© 2024. The Author(s).
Conflict of interest statement
J.-F.B., S.J.F.K., J.N., M.vL., and O.G. have been named inventor in a patent application claiming the discovery of this class of antiviral molecules as DENV replication inhibitors (WO 2017/167951), which was filed by applicants Janssen Pharmaceuticals, Inc. and Katholieke Universiteit Leuven, and has been granted in certain countries. O.G., J.-F.B., M.vL., S.J.F.K. and J.N. have been named inventors in a pending patent application relating to the use of substituted indole derivatives and substituted indoline derivatives in the manufacture of a medicament for the treatment or the prevention of dengue disease in an individual at risk of being infected by DENV and to a method for the treatment or the prevention of dengue in an individual at risk of being infected by DENV, which was filed by Applicants Janssen Pharmaceuticals, Inc. and Katholieke Universiteit Leuven (WO 2021/094563). O.G., B.G. and M.vL. are all full-time employees of Janssen and potential stockholders of Johnson and Johnson. The other authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

