IL-1R signaling drives enteric glia-macrophage interactions in colorectal cancer
- PMID: 39030280
- PMCID: PMC11271635
- DOI: 10.1038/s41467-024-50438-2
IL-1R signaling drives enteric glia-macrophage interactions in colorectal cancer
Abstract
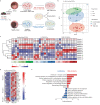
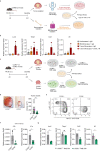
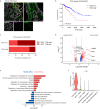
Enteric glia have been recently recognized as key components of the colonic tumor microenvironment indicating their potential role in colorectal cancer pathogenesis. Although enteric glia modulate immune responses in other intestinal diseases, their interaction with the colorectal cancer immune cell compartment remains unclear. Through a combination of single-cell and bulk RNA-sequencing, both in murine models and patients, here we find that enteric glia acquire an immunomodulatory phenotype by bi-directional communication with tumor-infiltrating monocytes. The latter direct a reactive enteric glial cell phenotypic and functional switch via glial IL-1R signaling. In turn, tumor glia promote monocyte differentiation towards pro-tumorigenic SPP1+ tumor-associated macrophages by IL-6 release. Enteric glia cell abundancy correlates with worse disease outcomes in preclinical models and colorectal cancer patients. Thereby, our study reveals a neuroimmune interaction between enteric glia and tumor-associated macrophages in the colorectal tumor microenvironment, providing insights into colorectal cancer pathogenesis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
- G0D8317N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- G0A7919N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- G086721N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- G088816N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- S008419N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- G067821N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- 11L0822N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- G067821N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- 1S44123N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- G0B4620N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)
- C12/15/016/KU Leuven (Katholieke Universiteit Leuven)
- C14/17/097/KU Leuven (Katholieke Universiteit Leuven)
- C3 grant, C3/21/037 or C3/23/067/KU Leuven (Katholieke Universiteit Leuven)
- F/2020/1512/Stichting Tegen Kanker (Belgian Foundation Against Cancer)
- F/2020/1512/Stichting Tegen Kanker (Belgian Foundation Against Cancer)
- 79756-GLIAMAC/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Skłodowska-Curie Actions (H2020 Excellent Science - Marie Skłodowska-Curie Actions)
- 833816-NEUMACS/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

