PD-1/LAG-3 co-signaling profiling uncovers CBL ubiquitin ligases as key immunotherapy targets
- PMID: 39030301
- PMCID: PMC11319776
- DOI: 10.1038/s44321-024-00098-y
PD-1/LAG-3 co-signaling profiling uncovers CBL ubiquitin ligases as key immunotherapy targets
Abstract
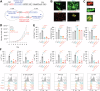
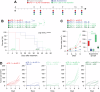
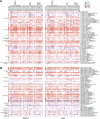
Many cancer patients do not benefit from PD-L1/PD-1 blockade immunotherapies. PD-1 and LAG-3 co-upregulation in T-cells is one of the major mechanisms of resistance by establishing a highly dysfunctional state in T-cells. To identify shared features associated to PD-1/LAG-3 dysfunctionality in human cancers and T-cells, multiomic expression profiles were obtained for all TCGA cancers immune infiltrates. A PD-1/LAG-3 dysfunctional signature was found which regulated immune, metabolic, genetic, and epigenetic pathways, but especially a reinforced negative regulation of the TCR signalosome. These results were validated in T-cell lines with constitutively active PD-1, LAG-3 pathways and their combination. A differential analysis of the proteome of PD-1/LAG-3 T-cells showed a specific enrichment in ubiquitin ligases participating in E3 ubiquitination pathways. PD-1/LAG-3 co-blockade inhibited CBL-B expression, while the use of a bispecific drug in clinical development also repressed C-CBL expression, which reverted T-cell dysfunctionality in lung cancer patients resistant to PD-L1/PD-1 blockade. The combination of CBL-B-specific small molecule inhibitors with anti-PD-1/anti-LAG-3 immunotherapies demonstrated notable therapeutic efficacy in models of lung cancer refractory to immunotherapies, overcoming PD-1/LAG-3 mediated resistance.
Keywords: CBL Ubiquitin Ligases; Cancer Immunotherapy; LAG-3; PD-1; T-cell Dysfunctionality.
© 2024. The Author(s).
Conflict of interest statement
CJE, JL and DE are inventors of the Humabody CB213 (WO/2019/158942. Crescendo Biologics Ltd.). The rest of the authors declare no competing interests.
Figures








References
-
- Arasanz H, Zuazo M, Bocanegra A, Gato M, Martinez-Aguillo M, Morilla I, Fernandez G, Hernandez B, Lopez P, Alberdi N et al (2020) Early detection of hyperprogressive disease in non-small cell lung cancer by monitoring of systemic T cell dynamics. Cancers 12:344 10.3390/cancers12020344 - DOI - PMC - PubMed
-
- Baraibar I, Roman M, Rodriguez-Remirez M, Lopez I, Vilalta A, Guruceaga E, Ecay M, Collantes M, Lozano T, Alignani D et al (2020) Id1 and PD-1 Combined Blockade impairs tumor growth and survival of KRAS-mutant lung cancer by stimulating PD-L1 expression and tumor infiltrating CD8(+) T cells. Cancers 12:3169 10.3390/cancers12113169 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- FIS PI20/00010/MEC | Instituto de Salud Carlos III (ISCIII)
- FIS PI23/00196/MEC | Instituto de Salud Carlos III (ISCIII)
- COV20/00237/MEC | Instituto de Salud Carlos III (ISCIII)
- FI21/00080/MEC | Instituto de Salud Carlos III (ISCIII)
- TRANSPOCART ICI19/00069/MEC | Instituto de Salud Carlos III (ISCIII)
- PFIS,FI21/00080/MEC | Instituto de Salud Carlos III (ISCIII)
- BMED 050-2019/Departamento de Salud, Gobierno de Navarra (Department of Health, Government of Navarra)
- BMED 51-2021/Departamento de Salud, Gobierno de Navarra (Department of Health, Government of Navarra)
- BMED 036-2023/Departamento de Salud, Gobierno de Navarra (Department of Health, Government of Navarra)
- PROYE16001ESC/Fundación Científica Asociación Española Contra el Cáncer (AECC)
- AGATA,0011-1411-2020-000013/Dirección General de Industria, Energia y Proyectos Estrategicos S3, Gobierno de Navarra (Department of Industry of the Government of Navarra)
- LINTERNA,0011-1411-2020-000033/Dirección General de Industria, Energia y Proyectos Estrategicos S3, Gobierno de Navarra (Department of Industry of the Government of Navarra)
- DESCARTHES,0011-1411-2019-000058/Dirección General de Industria, Energia y Proyectos Estrategicos S3, Gobierno de Navarra (Department of Industry of the Government of Navarra)
- ARNMUNE,0011-1411-2023-000101/Dirección General de Industria, Energia y Proyectos Estrategicos S3, Gobierno de Navarra (Department of Industry of the Government of Navarra)
- ISOLDA,grant agreement 848166/EC | Horizon 2020 Framework Programme (H2020)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

